Figure 3. Defective spindle orientation in cnn, Lis1, and dynactin (Gl) mutant neuroblasts.
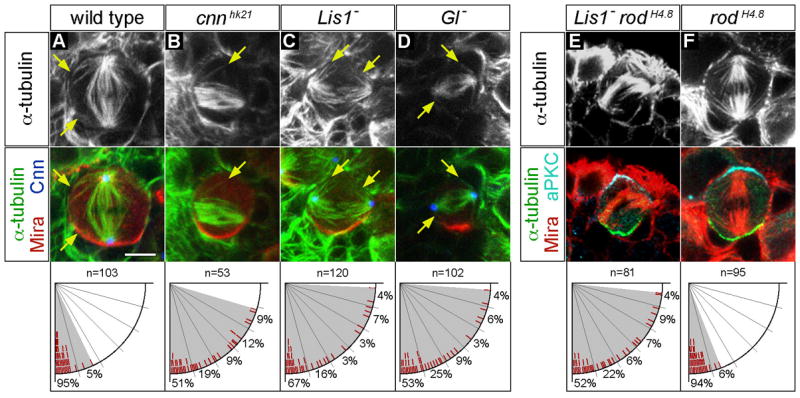
(A–D) wild type (A), cnnhk21 (B), Lis1 (C) and Gl (D) mutant neuroblasts were triple labelled for α-tubulin (top row), Cnn, and Mira (shown as merged images in bottom row).
(A) wild type metaphase neuroblasts showed spindles containing robust apical and basal asters nucleated by centrosomes (marked with Cnn). Note the proximity of astral MTs to the cortex (arrows) and the tight alignment of the spindle along the cell polarity axis (defined by basal Mira localization).
(B) cnnhk21 mutant neuroblasts lacked detectable Cnn protein, formed bipolar spindles with very few astral MTs, and often failed to align the spindle with the cell polarity axis.
(C–D) Lis1 (Lis1G10.14/Lis1k13209) and Gl (Gl1–3/Df(3L)fz-GF3b) mutant neuroblasts showed spindle alignment defects despite formation of bipolar spindles and astral microtubules (arrows) nucleated by centrosomes (marked with Cnn). The deficiency Df(3L)fz-GF3b removes the entire Gl locus.
(E–F) Lis1 rodH4.8 double and rodH4.8 single mutant neuroblasts were triple labelled for α-tubulin (top row), aPKC, and Mira (shown as merged images in bottom row).
(E) Lis1 rodH4.8 double mutant metaphase neuroblasts showed spindle alignment defects as observed in Lis1 single mutants.
(F) Metaphase spindle orientation is normal in rodH4.8 single mutant neuroblasts. Scale bar: 5 μm.
