Fig. 5. Electrophoretic mobility shift analysis of proteins interacting specifically with the mouse or human 5′ upstream DYT1 region.
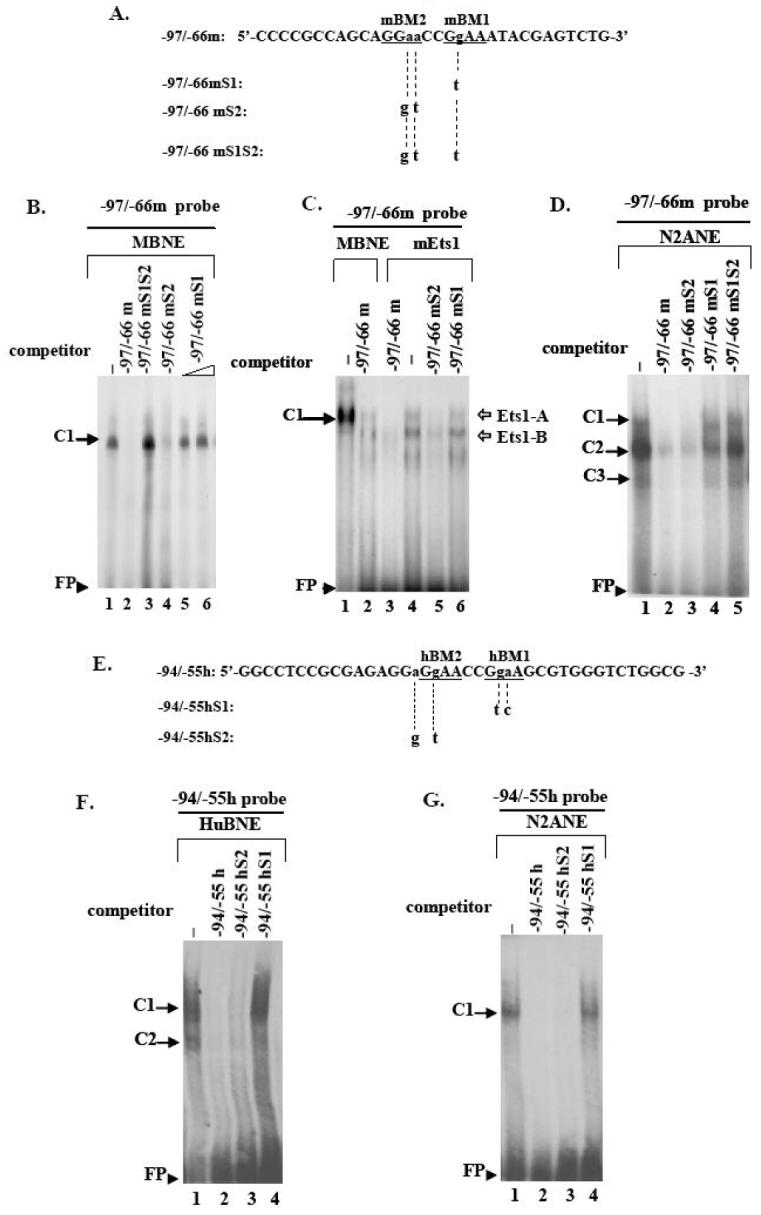
(A, E) Coding strand nucleotide sequence of mouse -97/-66 (-97/-66m, A) and human -94/-55 wildtype probes (-94/-55h, E) used in EMSAs. Underlined bases indicate examined Ets binding sites; the -86bp to -83bp and -80bp to -77bp Ets mouse cores have been designated as mEts2 and mEts1, respectively (A) while the -78bp to -75bp and -72bp to -69bp human Ets cores have been designated as hEts2 and hEts1, respectively (E). Small letters indicate bases substituted to produce the mouse mutant versions -97/-66mS1, -97/-66mS2, -97/-66mS1S2 (A) or the human mutant versions -94/-55hS1, -94/-55hS2 (E), for which only the altered bases are shown. Substitutions are also shown in Fig. 4. (A2). A retarded DNA-protein complex C1 was formed after incubation of mouse brain nuclear extract (MBNE) with the -97/-66m probe in the absence of competitor (lane 1), which was abrogated in the presence of 100-fold excess of homologous -97/-66m (lane 2) or heterologous -97/-66mS2 (lane 4) competitor. C1 was retained in the presence of 100-fold excess of -97/-66mS1S2 (lane3) or 100 to 200-fold excess of -97/-66mS1 (lanes 5, 6) competitor. (A3). Incubation of in vitro translated mouse Ets1 protein (mEts1) with the -97/-66m probe led to two retarded DNA-mEts1 complexes (Ets1-A and Ets1-B) (lane 4). Ets1-A complex exhibits the same retardation velocity as the C1 complex resulting from interaction of the -97/-66m probe with mouse nuclear extract (lane 1). Competition with 100-fold excess of either homologous (lane 3) or heterologous -97/-66mS2 (lane 5) competitor abolishes both complexes, while competition with -97/-66mS1 probe does not (lane 6). (B2). Human brain nuclear extract (HuBNE) incubated with the -94/-55h probe in absence of competitor gave rise to two DNA-protein complexes C1, C2 (lane 2), both of which were abolished in the presence of 100-fold excess of homologous -94/-55h (lane 1), or heterologous -94/-55hS2 (lane 3) competitor, while they were retained intact when competed with 100 fold excess of -94/-55hS1 probe (lane 4). DNA-nuclear extract protein retarded complexes are indicated by black arrows while DNA-mEts1 protein complexes by white arrows. Free probe is indicated by arrowheads. Negative controls in the absence of nuclear extract or mEts1 protein did not produce any specific binding (data not shown).
