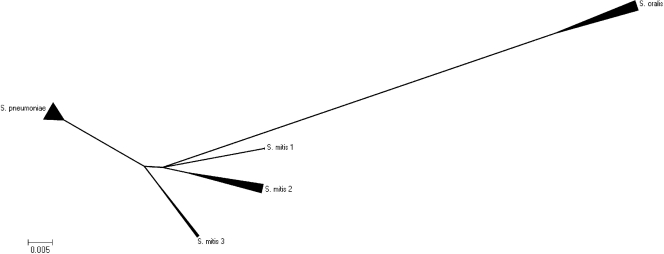
FIG. 2.
Minimal evolution algorithm (suppressed) obtained by using the MEGA (version 4.0) program and based on the partial gdh gene sequences of 11 S. oralis strains, 17 S. pneumoniae strains, and 13 S. mitis strains. It shows that the three species form three distinct clusters. The S. oralis cluster has a longer distance to the two other clusters, indicating that S. pneumoniae and S. mitis are genetically more closely related on the basis of gdh gene evolution. There are three subclusters within the S. mitis cluster, indicating that the species S. mitis contains a heterogeneous group of strains.