Abstract
The human gastric pathogen Helicobacter pylori has many virulence factors involved in pathogenesis, but the mechanisms regulating these virulence factors are not yet fully understood. In this study, we cloned HP1248, which is similar in sequence to Escherichia coli vacB, which was previously shown to be associated with the expression of virulence in Shigella and enteroinvasive E. coli. E. coli vacB encodes RNase R. RNase R is involved in the posttranscriptional regulation of mRNA stability. By global transcriptional microarray profiling of an H. pylori HP1248 deletion mutant, we defined six virulence-related genes which were posttranscriptionally downregulated by HP1248, including the motility-related genes HP1192 and flaB, the chemotaxis-related gene cheY, and the apoptosis-inducing genes HP0175, cagA, and gtt. In this study, recombinant HP1248 protein expressed in E. coli showed 3′-to-5′ exoribonuclease activity. Motility and apoptosis induction were increased in the H. pylori HP1248 deletion mutant. We also showed that HP1192 is associated with H. pylori motility, possibly through HP1248 regulation. Further, we suggested and studied the possible mechanisms of this specific regulation of virulent genes by HP1248. In addition, the expression level of HP1248 mRNA changed dramatically in response to a variety of altered environmental conditions, including pH and temperature. Hence, HP1248 in H. pylori seems to play a role in environmental sensing and in regulation of virulent phenotypes, such as motility and host apoptosis induction.
Helicobacter pylori (27) is an important human pathogen, responsible for type B gastritis and peptic ulcers and for increasing the risk of gastric adenocarcinoma and mucosa-associated lymphoid tissue lymphoma of the stomach (6, 32, 35, 42).
Several bacterial factors, including flagella, various enzymes, and toxins, contribute to the full virulence of H. pylori (5, 9, 13, 14, 33). The motility of H. pylori is provided by flagella, the filaments of which consist of two flagellin types (38). The majority of the filament is composed of FlaA and FlaB (24). Motility in H. pylori is essential for colonization in gnotobiotic piglets (15). H. pylori infection can also induce apoptosis in gastric epithelial cells, lymphocytes, and macrophages (17, 31, 45) and could contribute to mucosal inflammation. In addition, the loss of activated macrophages is likely to decrease the effective immune response to the pathogen (17). Recently some apoptosis-inducing factors of H. pylori were examined (16, 25). However, the regulation mechanism of these virulence factors is not yet fully understood.
In this study, we used an expression library of H. pylori (10) to clone a possible chlorhexidine resistance-related gene, HP1248. However, we suggested that the chlorhexidine-resistant clone might be an overexpression artifact. On the other hand, HP1248 is similar to Escherichia coli vacB based on sequence similarity. Although its exact mechanism of action is unknown, vacB has been shown to encode RNase R and to be associated with the expression of virulence in Shigella and enteroinvasive E. coli (12).
RNase R is a 3′-5′ exoribonuclease that has homologues widespread in most sequenced genomes. E. coli RNase R participates in RNA quality control and is involved in the surveillance of stable RNAs (26). In the absence of RNase R, the small stable SsrA/tmRNA is not properly processed (8, 20). RNase R might have a role in the decay of structured mRNA (11); mRNA decay is important for determining levels of gene expression. In addition, RNase R is involved in the posttranscriptional regulation of mRNA stability by 3′-to-5′ exonucleolytic degradation (3, 40).
So, we hypothesized that HP1248 might encode RNase R and that mRNA decay induced by HP1248 RNase activity might be one of the mechanisms regulating virulence factor mRNAs in H. pylori. One purpose of this study was to determine which genes are upregulated in an H. pylori HP1248 deletion mutant. We showed that several virulence-related genes were posttranscriptionally downregulated by HP1248, including HP0175, gtt, cagA, flaB, cheY, and HP1192. HP0175, cagA, and gtt can induce apoptosis of host cells (5, 23, 30, 39). In addition, flaB is involved in the motility of H. pylori (37) and HP1192 is a putative motility protein. In H. pylori, cheY is involved in chemotaxis (41). We also found and suggested the possible mechanism of this specific regulation of virulent genes by HP1248. These data suggested that the regulation of virulence factors by HP1248 may play an important role in helping cells cope with a changing environment.
MATERIALS AND METHODS
Bacterial strains and growth conditions.
Clinical isolates were obtained at the National Taiwan University Hospital (NTUH) as described previously (19). H. pylori strains were grown on Columbia blood agar plates containing 5% sheep blood and chloramphenicol (4 μg/ml), kanamycin (10 μg/ml), or chlorhexidine (3 μg/ml) and incubated for 2 to 3 days under microaerophilic conditions (5% O2, 10% CO2, 85% N2) at 37°C. For liquid culture, H. pylori bacteria were grown in a Brucella broth containing 5% fetal calf serum. The pH value of the medium was titrated by using HCl or NaOH. For experiments under various stress conditions, cells growing in Brucella broth (pH 7.0) at 37°C were rapidly transferred at mid-exponential phase to 22 to 30°C or acid Brucella broth (pH 4.0 to pH 5.5) and then incubated for 4 h under microaerophilic conditions.
Escherichia coli strains were grown on LB agar plates or in LB broth containing appropriate antibiotics or chlorhexidine.
Construction of H. pylori deletion mutants and overexpression mutant and complementation.
For construction of the HP1248 deletion mutant and HP1192 deletion mutant, the genes and flanking region which encode HP1248 and HP1192 were amplified from the genomic DNA of a wild-type NTUH-C1 strain by PCR and then cloned into a pGEM-T Easy plasmid (Promega). The HP1248 deletion construct was generated by inverse PCR using the HP1248-Inverse F (5′-CAAAAAACACTTCGCATGACTTC-3′) and HP1248-Inverse R (5′-AGATTTGGACCATTATTTAAAAC-3′) primers. The primers HP1192-Inverse F (5′-AAACAGAGAAAACATGATAACG-3′) and HP1192-Inverse R (5′-CGCTTTTTACTCTTAAAATTTC-3′) were used to generate the HP1192 deletion construct. A blunt-end PCR product of the cat coding region was amplified by a Pfu polymerase using the CAT(+1)F (5′-ATGCAATTCACAAAGATTG-3′) and CAT(+624)R (5′-TTATTTATTCAGCAAGTC-3′) primers and then phosphorylated by a polynucleotide kinase (New England Biolabs, Beverly, MA). A blunt-end PCR product of the km coding region was amplified by a Pfu polymerase using the KM(+1)F (5′-ATGGCTAAAATGAGAATATCACC-3′) and KM(+795)R (5′-CTAAAACAATTGATCCAG-3′) primers by the same method. The inverse PCR products and blunt-end cat or blunt-end km gene were ligated. The orientation of the cat or km gene was the same as the direction of the operon. We obtained two plasmids in which HP1248 and HP1192 were replaced by a cat coding region and a km coding region, respectively. These plasmids were naturally transformed into the wild type to generate the C1/HP1248-cat deletion mutant and the C1/HP1192-km deletion mutant such that the insertion site of the cat or km gene is at nucleotide 1. By transforming into the C1/HP1192-km deletion mutant the plasmid in which HP1248 was replaced by a cat coding region, we obtained the C1/HP1248-cat/HP1192-km double mutant.
For overexpression of HP1192 and flaB, PCR was performed to amplify the predicted promoter regions of flaA corresponding to positions −164 to +1 relative to the transcriptional start site. This predicted promoter of the flaA gene is named flaA′ and is an active promoter in H. pylori (7). Then, the predicted promoter region, flaA′, was introduced upstream of flaB or HP1192 (SacII site of pGEM-T easy/flaB; SacII site of pGEM-T easy/HP1192). The resulting flaA′-flaB and flaA′-HP1192 constructs were cloned into the XhoI site of pHel3 (18).
For complementation of the C1/HP1248-cat deletion mutant, the PCR product of HP1248 containing the predicted promoter region was cloned into the single EcoRV site of pHel3 (18), and then the PCR product of HP1248 containing the predicted promoter region and the kanamycin resistance gene was cloned into the single HindIII site of the pGEM-T Easy/HP0405 plasmid. HP0405 encodes a NifS-like protein, and the inactivation of this gene did not affect the growth rate (1). The complementation construct was naturally transformed into the C1/HP1248-cat deletion mutant. The gene alignments of these complementation strains were confirmed by PCR using different combinations of primers.
Microarray hybridization and data analysis.
Microarrays of H. pylori strain 26695, which were spotted in four repeats, were kindly provided by The Institute of Genome Research (http://www.tigr.org). Total RNA of H. pylori strains was extracted as described previously (4). To determine which genes are differentially expressed in the C1/HP1248-cat mutant, cDNA was prepared from RNA extracted from the H. pylori NTUH-C1 and C1/HP1248-cat strains and was hybridized to microarray slides. The RNA samples from two independent preparations from strains NTUH-C1 and C1/HP1248-cat, respectively, were used for cDNA labeling and were hybridized to two independent microarray slides. Each slide contained four set of probe for the same open reading frame in strain 26695. Equal amounts of RNA (20 μg) were used to synthesize differentially labeled cDNA (Cy3-dCTP and Cy5-dCTP; Amersham Biosciences) during first-strand reverse transcription reactions with Superscript II reverse transcriptase and 6 μg of random primers (Invitrogen). Synthesized cDNAs were purified by using S.N.A.P. columns (Invitrogen), and hybridizations were performed according to the manufacturer's protocol. After being washed, the slides were scanned using a ScanArray HT scanner and analyzed by using the ScanArray Express software program (Perkin Elmer). Spots were flagged and eliminated from analysis when the signal-to-background ratio was less than 3 or in obvious instances of high background or stray fluorescent signals. Median intensities of spots were background corrected, and differences in dye bias were normalized by using the LOWESS algorithm. To determine the significance of differential expression, the signal ratios used as measures of differential expression between the red and green channels were obtained from processed signal intensities. The value in the ratio column represents the average of eight sets of signal ratios of those genes (Tables 1 and 2). Signal ratios of >2.0 or <0.5 were analyzed further.
TABLE 1.
HP1248-repressed genesa
| ORF no. | Genome organizationb | HP1248−/WTc ratiod | Putative gene product,e gene |
|---|---|---|---|
| HP0031 | op HP0031-HP0037 | 2.16 ± 0.88 | Hypothetical protein |
| HP0114 | op HP0114-HP0115 | 2.01 ± 0.31 | Hypothetical protein |
| HP0115 | op HP0114-HP0115 | 3.14 ± 0.29 | Flagellin B, flaB |
| HP0130 | m | 3.07 ± 0.70 | Hypothetical protein |
| HP0175 | op HP0175-HP0177 | 2.19 ± 0.23 | Cell binding factor 2 |
| HP0176 | op HP0175-HP0177 | 2.08 ± 0.44 | Fructose-bisphosphate aldolase, tsr |
| HP0194 | op HP0194-HP0196 | 2.28 ± 0.53 | Triosephosphate isomerase, tpi |
| HP0231 | op HP0231-HP0234 | 2.32 ± 0.61 | Hypothetical protein |
| HP0390 | m | 2.49 ± 0.23 | Adhesin-thiol peroxidase, tagD |
| HP0486 | op HP0486-HP0487 | 2.61 ± 0.28 | Hypothetical protein |
| HP0547 | op HP0547-HP0548 | 2.35 ± 0.24 | cag pathogenicity island protein, cagA |
| HP0631 | op HP0631-HP0635 | 2.23 ± 0.36 | Quinone-reactive Ni/Fe, hydrogenase, small subunit, hydA |
| HP0653 | op HP0653-HP0651 | 2.01 ± 0.66 | Nonheme iron-containing ferritin, pfr |
| HP0671 | m | 2.97 ± 0.49 | Outer membrane protein, omp14 |
| HP0682 | op HP0682-HP0681 | 2.22 ± 0.81 | Hypothetical protein |
| HP0706 | m | 2.32 ± 0.52 | Outer membrane protein, omp15 |
| HP0795 | op HP0796-HP0795 | 2.13 ± 0.67 | Trigger factor, tig |
| HP0950 | op HP0950-HP0948 | 2.07 ± 0.47 | Acetyl-CoA carboxylase beta subunit, accD |
| HP1067 | op HP1067-HP1069 | 2.04 ± 0.19 | Chemotaxis protein, cheY |
| HP1083 | op HP1083-HP1080 | 3.73 ± 0.33 | Hypothetical protein |
| HP1118 | m | 2.02 ± 0.10 | Gamma-glutamyltranspeptidase, ggt |
| HP1124 | op HP1129-HP1123 | 3.98 ± 0.96 | Hypothetical protein |
| HP1137 | op HP1137-HP1131 | 2.17 ± 0.48 | ATP synthase F0, subunit b′, atpF′ |
| HP1151 | op HP1153-HP1147 | 2.05 ± 0.78 | Ribosomal protein S16, rps16 |
| HP1192 | m | 2.98 ± 0.28 | Secreted protein involved in flagellar motility |
| HP1288 | op HP1288-HP1289 | 2.82 ± 0.39 | Hypothetical protein |
| HP1454 | op HP1457-HP1454 | 2.14 ± 0.77 | Hypothetical protein |
Genes listed are those whose transcription differed more than twofold (ratio > 2.0) for the HP1248 deletion mutant H. pylori C1/HP1248-cat compared to that for the NTUH-C1 wild type, according to microarray analysis. ORF, open reading frame.
m, monocistronically transcribed genes; op, putative transcription unit.
WT, wild type. HP1248−, HP1248 deletion mutant.
Ratios are averages ± standard deviations.
The functional annotation is that used by the TIGR database (http://www.tigr.org). CoA, coenzyme A.
TABLE 2.
HP1248-induced genesa
| ORF no. | HP1248−/WTb ratio | Putative gene product,c gene |
|---|---|---|
| HP1177 | 0.48 | Outer membrane protein, omp27 |
| HP0606 | 0.49 | Membrane fusion protein, mtrC |
| HP0229 | 0.25 | Outer membrane protein, omp6 |
| HP0298 | 0.48 | Dipeptide ABC transporter, dppA |
| HP0299 | 0.44 | Dipeptide ABC transporter, dppB |
| HP1340 | 0.40 | Biopolymer transport protein, exbD |
| HP0687 | 0.46 | Iron(II) transport protein, feoB |
| HP0514 | 0.49 | Ribosomal protein L9, rpl9 |
| HP0691 | 0.46 | 3-Oxoadipate CoA-transferase subunit A, yxjD |
| HP1302 | 0.49 | Ribosomal protein S5, rps5 |
| HP1300 | 0.25 | Preprotein translocase subunit, secY |
| HP1256 | 0.48 | Ribosome releasing factor, frr |
| HP0215 | 0.49 | CDP-diglyceride synthetase, cdsA |
| HP1237 | 0.46 | Carbamoyl-phosphate synthetase, pyrAa |
| HP0399 | 0.48 | Ribosomal protein S1, rps1 |
| HP1186 | 0.15 | Carbonic anhydrase |
| HP1555 | 0.43 | Translation elongation factor EF-Ts, tsf |
| HP0306 | 0.48 | Glutamate-1-semialdehyde 2,1-aminomutase, hemL |
| HP0690 | 0.48 | Acetyl CoA acetyltransferase (thiolase), fadA |
| HP1299 | 0.25 | Methionine amino peptidase, map |
| HP1239 | 0.49 | Hypothetical protein |
Genes listed are those whose transcription differed less than half (ratio < 0.5) for the HP1248 deletion mutant H. pylori C1/HP1248-cat compared to that for the NTUH-C1 wild type, according to microarray analysis. ORF, open reading frame.
WT, wild type. HP1248−, HP1248 deletion mutant.
The functional annotation is that used by the TIGR database (http://www.tigr.org). CoA, coenzyme A.
Slot blot.
Slot blot and probe preparation were performed as described previously (44). Densitometry was analyzed using the NIH Image version 1.62 software program by using ureA or 23S rRNA as the internal standard. Although ureA is inducible in acidic pH, the ureA transcript was chosen as the internal control because it was expressed consistently across different genotypes at the same growth phase in the same pH (36). Because environmental conditions could affect ureA transcription, for these assays the 23S rRNA gene was chosen for normalization. When we used 23S rRNA as the internal standard, the slot blot was performed with 1 μg of total RNA from each H. pylori isolate to avoid overexposure of the films.
Expression and purification of protein.
The gene encoding the HP1248 protein was amplified from the DNA of a wild-type NTUH-C1 strain by PCR and cloned into a TA vector, pGEM-T Easy, to create pGEM-T Easy/HP1248. In order to generate the HP1248 D231N mutant, pGEM-T Easy/D231N, site-directed mutagenesis was performed using the D231N-F (5′-CCCAAAGACGCTAAAGATTTTAACGATGCGATTTTTTATGACC-3′) primer and pGEM-T Easy/HP1248 according to the manufacturer's instructions (QuikChange II site-directed mutagenesis kit; Qiagen). The pGEM-T Easy/HP1248 and pGEM-T Easy/D231N plasmids were digested by SalI and BamHI (New England Biolabs, Beverly, MA) and ligated in frame into the pET28a plasmid (Novagen, Darmstadt, Germany). The resulting pET28a/HP1248 plasmid and pET28a/D231N plasmid were transformed into E. coli strain BL21(DE3). The HP1248 protein and D231N mutant protein were expressed and purified per the manufacturer's instruction under 1 mM isopropyl-1-thio-β-d-galactopyranoside induction at 25°C (Qiagen, Hilden, Germany).
Nuclease activity assay.
The templates for HP1192 were synthesized by PCR using the primers HP1192-F (5′-ATGAAAAAGCAAATCTTGACAG-3′) and HP1192-R (5′-TTAGACCACTGAGTTTTTAGG-3′) and cloned into a TA vector, pGEM-T Easy. Uniformly labeled RNA substrates for the RNase assay were synthesized by T7 RNA polymerase transcription (MAXIscript T7 kit; Ambion) in the presence of [α-32P]UTP. We used polynucleotide kinase (Biolabs) and [γ-32P]ATP (GE-Amersham) to 5′ label the nonradioactive transcripts. To perform 3′ labeling, we used RNA ligase (Biolabs) and [32P]pCp (GE-Amersham) for 6 h at 18°C in the presence of 10% dimethylsulfoxide.
The HP1248 protein was assayed in a 50-μl reaction volume containing 20 mM Tris-HCl at pH 8.0, 0.25 mM MgCl2, 300 mM KCl, and 0.1 mM dithiothreitol. The amounts of enzyme varied from 0.05 to 0.1 mg, and the amount of substrate varied from 0.2 pmol to 1.0 pmol, as indicated. Reactions were incubated at 25°C for the times indicated, stopped by addition of 5 ml of 95% formamide, 20 mM EDTA, 0.05% bromophenol blue, and 0.05% xylene cyanol, and loaded on 20% polyacrylamide-7 M urea gels.
Motility assay.
To test the motility of H. pylori, 107 bacteria were spotted on a 4-mm-diameter sterilized filter paper and then the filter paper was put on plates of Brucella medium with 0.4% agar and 10% (vol/vol) fetal bovine serum (FBS). The plates were incubated at 37°C for 4 days under microaerophilic conditions, and motility was assessed by examining the size of the swarming halo.
Eukaryotic-cell culture and enzyme-linked immunosorbent assay (ELISA) apoptosis analysis.
The murine macrophage cell line RAW 264.7 was maintained in Dulbecco's modified Eagle medium supplemented with 10% FBS and 1% penicillin-streptomycin at 37°C in a humidified 5%-CO2 atmosphere. For the coculture experiments, RAW 264.7 cells were plated in the same medium without penicillin-streptomycin for 2 h, and H. pylori was then added to the macrophages at a multiplicity of infection (MOI) of 30:1 or 50:1. A previous study (30) showed that the level of apoptosis was significant at an MOI of 50:1 but not at 30:1. Therefore, we used MOI of 30:1 and 50:1 for comparison. RAW 264.7 cells (2 × 105 cells/well) were cultured in 24-well plates in the presence or absence of H. pylori or 0.2 ppm staurosporine as a positive control. Floating and adherent cells were harvested after the experiments and counted with a hemocytometer, and 104 cells were analyzed using the Cell Death Detection ELISA Plus kit from Roche Molecular Biochemicals (Indianapolis, IN), based on the determination of cytoplasmic histone-associated DNA fragments. According to the manufacturer's instructions, in each experiment the optical density (OD) of unstimulated control cells (medium only) was assigned a value of 1.0, and the relative amount of apoptosis in experimental groups was determined as a ratio to the control level.
Construction of xylE reporter genes and measurement of XylE activity.
Reporter genes were constructed with a promoterless Pseudomonas putida catechol-2,3-dioxygenase (xylE) reporter (46) and cloned into a TA vector, pGEM-T Easy. For construction of the flaA′-xylE, flaB′-xylE, cheY′-xylE, HP1192′-xylE, gtt′-xylE, cagA′-xylE, and HP0175′-xylE reporter genes, PCR was performed to amplify the predicted promoter regions corresponding to positions −164 to +1, −166 to +1, −291 to +1, −218 to +1, −211 to +1, −834 to +1, and −431 to +1 relative to the transcriptional start sites of these genes, respectively. We defined the promoter region of the gene that locates the noncoding region between two adjacent operons. Then, these predicted promoter regions were introduced upstream of xylE (SacII site of pGEM-T Easy/xylE). The resulting flaA′-xylE, flaB′-xylE, cheY′-xylE, HP1192′-xylE, gtt′-xylE, cagA′-xylE, and HP0175′-xylE reporter genes were cloned into the XhoI site of pHel3 (18).
Construction of pHel3/flaA′-xylE with the specific RNase recognition (RR) sequence was performed using the XYLE-RR-F (5′-CCGCAACGAAGTGTTGTGAGTGGGAGATTACTCACACCCGGACCAC-3′) primer according to the manufacturer's instructions (QuikChange II site-directed mutagenesis kit; Qiagen) to create pHel3/flaA′-xylE+RR. In addition, we complemented HP1248 or the D231N mutant with the shuttle vector pHel3/flaA′-xylE+RR; HP1248 or D231N, containing the predicted promoter region, was cloned into the SphI site of the pHel3/flaA′-xylE+RR vector. Then, the complementation shuttle vector was naturally transformed into the C1/HP1248-cat deletion mutant.
XylE activities in whole cells were measured as described previously (43). H. pylori strains containing the xylE reporter plasmids were grown on Columbia blood agar supplemented with kanamycin for 48 h and then resuspended in 50 mM phosphate buffer, pH 7.4, to a cell density of 1 OD at 600 nm unit, which corresponded to 109 CFU/ml. Reactions were initiated by adding cells (50 to 100 μl) to reaction mixtures containing 10 mM catechol in 50 mM potassium phosphate, pH 7.4. Catechol oxidation to 2-hydroxymuconic semialdehyde was monitored continuously at 375 nm using a Beckman DU 640B recording spectrophotometer at room temperature. A unit of XylE activity corresponds to 1 μmol of catechol/min, and values were expressed as units per minute per 108 cells.
RESULTS
Cloning of HP1248 gene.
Screening of the phagemid expression library in XLOLR (10) revealed three clones that grew on plates containing 3 μg/ml chlorhexidine, whereas E. coli strain XLOLR with phagemid only did not. DNA sequencing of the three clones revealed the same gene locus. The sequences revealed a single large open reading frame (GenBank identifier AAD08293.1). A DNA homology search indicated that this open reading frame was H. pylori HP1248, and the sequence revealed 55.4% similarity and 36.0% identity at the nucleotide level to the vacB gene of E. coli (http://cmr.jcvi.org/cgi-bin/CMR/CmrHomePage.cgi).
To confirm that HP1248 was related to resistance, a chlorhexidine susceptibility test of the C1/HP1248-cat deletion mutant was performed, but the chlorhexidine susceptibility of the C1/HP1248-cat deletion mutant was the same as that of the wild-type strain. These results revealed that the chlorhexidine-resistant clone might be an overexpression artifact.
Expression of H. pylori genes on microarray.
Because the vacB gene has been previously shown to be associated with the expression of virulence in Shigella and enteroinvasive E. coli, we wanted to define what genes were controlled by HP1248, especially virulence-related genes. RNA expression of H. pylori NTUH-C1 and the C1/HP1248-cat deletion mutant were compared by microarray analysis. According to the growth curve assay, the growth rates of the wild-type and mutant strains were similar (data not shown). A total of 27 genes were identified as upregulated in the C1/HP1248-cat deletion mutant by using a signal ratio cutoff of 2.0 (Table 1). In addition, a total of 21 genes were identified as downregulated in the C1/HP1248-cat deletion mutant by using a signal ratio cutoff of 0.5 (Table 2). Because we proposed that HP1248 might encode an exoribonuclease and downregulate genes by mRNA decay, we first focused on genes which were upregulated in the C1/HP1248-cat deletion mutant. A number of virulence-related genes involved in pathogenesis were upregulated. Three genes, including flaB (37), HP1192, and cheY (41), encode motility- and chemotaxis-related proteins; the flaB gene and HP1192 were upregulated about threefold. Three genes, HP0175, cagA, and gtt, encode apoptosis-related proteins (5, 23, 30, 39). Results showed some operons involved and some genes are monocistronically transcribed genes (Table 1).
Confirmation of expression levels in slot blotting and microarray analysis.
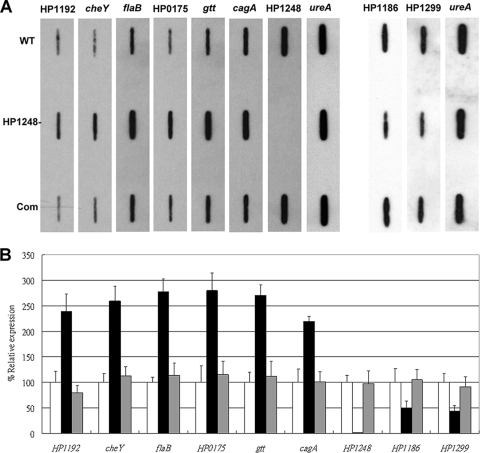
To confirm the differential expression of the genes identified by microarray analysis, six virulence-related genes, including HP0175, gtt, cagA, flaB, cheY, and HP1192, were selected and their RNA expressions were monitored by RNA slot blot analysis in the NTUH-C1 wild type, the C1/HP1248-cat deletion mutant, and the C1/HP1248-cat deletion mutant complemented with HP1248 in HP0405. On the other hand, two HP1248-induced genes identified by microarray analysis, HP1186 and HP1299, were also selected, and their RNA expressions were monitored by RNA slot blot analysis. The results shown in Fig. 1A and B agreed with those from the microarray analysis; the RNA levels of HP0175, gtt, cagA, flaB, cheY, and HP1192 were found to be increased in the C1/HP1248-cat deletion mutant. Complementation with HP1248 restored the RNA levels of HP0175, gtt, cagA, flaB, cheY, HP1192, HP1186, and HP1299 to the levels for the NTUH-C1 wild type. The ureA transcript was chosen for an internal control because it was expressed consistently across different genotypes at the same growth phase.
FIG. 1.
(A) Analysis of RNA expression of six selected virulence genes and two HP1248-induced genes by slot blot hybridization. The amount of RNA used for all conditions was standardized at 10 μg per slot. RNA extracted from H. pylori NTUH-C1 (wild type [WT]), the C1/HP1248-cat mutant (HP1248−), and the complementation strain (Com) was used in the respective experiments performed with probes specific for the indicated genes. RNA slot blot analysis with labeled probes specific for ureA and HP1248 were performed as a control. (B) Quantitative analysis of triplicate RNA expression normalized with ureA RNA expression. The expression level of each gene for the WT (white bars) was assigned a value of 100, and the relative level of RNA expression for HP1248− (black bars) or Com (gray bars) was determined as a ratio to that of the WT.
Involvement of HP1248 in posttranscriptional level of expression of virulence genes.
To determine whether upstream activation sequences are required for regulation of HP0175, gtt, cagA, flaB, cheY, and HP1192 by HP1248, we constructed pHel3 (18) carrying the predicted promoter region of HP0175, gtt, cagA, flaB, cheY, or HP1192 fused with the xylE reporter gene and then introduced them into the H. pylori strains NTUH-C1 and the C1/HP1248-cat deletion mutant. We also integrated cagA′-xylE in chromosomes (HP0405) of NTUH-C1 and the C1/HP1248-cat deletion mutant. The XylE activity expressed in each strain was measured at pH 7, and the results are shown in Table 3. The activity for each promoter was not reduced markedly in the mutant compared with that in the wild type. On the other hand, we also detected the XylE activity of cagA′-xylE on the wild type at pH 5. The cagA promoter was inducible under acid exposure. We can see that the XylE activity of cagA′-xylE was inducible at pH 5. The result strengthen the negative result of no difference in reporter activity between the HP1248 mutant and the wild type. Thus, these results indicated that gene regulation by HP1248 was not through transcription but might involve regulation at a posttranscriptional level for HP0175, gtt, cagA, flaB, cheY, and HP1192.
TABLE 3.
Promoter activities in the wild type and HP1248 deletion mutant
| H. pylori strain | Relevant genotype, pHa | XylE activity (U/min/108 cells) for reporter geneb
|
||||||
|---|---|---|---|---|---|---|---|---|
| cagA′-xylE | cheY′-xylE | HP1192′-xylE | flaB′-xylE | HP0175′-xylE | ggt′-xylE | cagA′-xylE(C)c | ||
| NTUH-C1 | WT, 7 | 12.6 ± 4.0 | 8.2 ± 3.3 | 7.1 ± 2.3 | 10.3 ± 3.1 | 6.6 ± 2.2 | 11.0 ± 3.8 | 2.3 ± 1.0 |
| C1/HP1248-cat | HP1248−, 7 | 13.4 ± 3.9 | 7.5 ± 3.8 | 7.7 ± 3.4 | 11.1 ± 4.9 | 5.9 ± 1.9 | 10.5 ± 2.9 | 1.9 ± 0.7 |
| NTUH-C1 | WT, 5 | 19.8 ± 3.1 | NAd | NA | NA | NA | NA | NA |
WT, wild type; HP1248−, HP1248 deletion.
Values represent the averages for three replicates ± standard deviations.
cagA′-xylE(C), cagA′-xylE integrated in chromosome HP0405 locus.
NA, not available.
HP1248 is a 3′-to-5′ exoribonuclease.
Exonucleases, unlike endonucleases, produce only mononucleotide products. Because HP1192 is a gene (282 nucleotides [nt] in length) among the upregulated virulence-related genes of the C1/HP1248-cat deletion mutant, it could be a good substrate of the HP1248 protein. We therefore synthesized uniformly labeled HP1192 RNA using [α-32P]UTP and performed an analysis of its degradation with purified HP1248 protein. The reaction mixtures were loaded directly on 20% denaturing polyacrylamide gels, permitting the detection of mononucleotides (34). In addition, E. coli RNase R was used as a positive control. As shown in Fig. 2, when the products of HP1248 degradation were separated on a 20% denaturing polyacrylamide gel, only the mononucleotide was detected after treatment with the HP1248 protein. Thus, we infer that the activity is exoribonucleolytic. The HP1248 D231N protein bearing a mutation (Asp231 to Asn) in its active site that inactivates the enzyme (2) had no exoribonuclease activity.
FIG. 2.
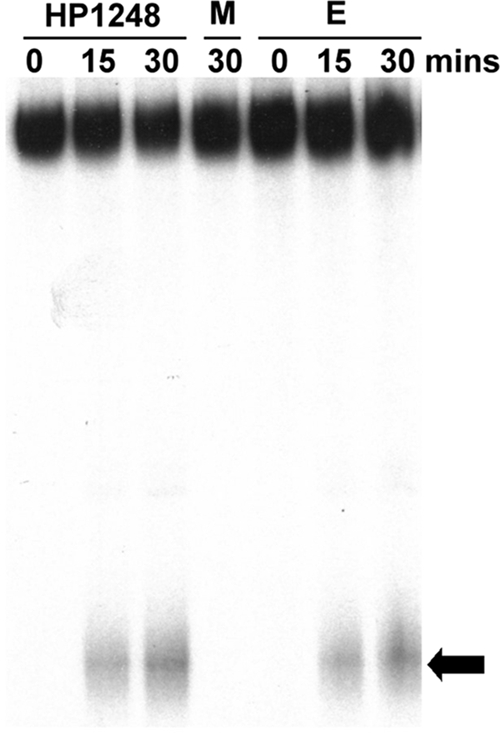
Exoribonuclease activity assay of the HP1248 protein. A total of 1.0 pmol labeled HP1192 RNA was incubated with 0.05 mg HP1248 for the times indicated and then separated by 20% denaturing polyacrylamide gel electrophoresis. The control reactions were carried out with the HP1248 protein D231N mutant (M) for 30 min and E. coli RNase R (E) for the times indicated. The arrow at right indicates labeled mononucleotide product.
To differentiate between 5′-to-3′ versus 3′-to-5′ exonucleolytic activity, we examined the degradation of a 20-nt synthetic RNA (pUGGUGGUGGAUCCCGGGAUC) (28) labeled at either its 5′ ([γ-32P]ATP) or 3′ ([α-32P]UTP) extremity, purified, and incubated for different times in the presence of the HP1248 protein. Digestion of the 3′-labeled RNA species by the HP1248 protein resulted in the accumulation of single product (Fig. 3A), while the D231N mutant had no exonucleolytic degradation. Digestion of the 5′-labeled RNA species, on the other hand, resulted in the production of many intermediate products (Fig. 3B), consistent with exonucleolytic attack from the 3′ end, while the D231N mutant had no exonucleolytic degradation. Thus, we conclude that HP1248 has a 3′-to-5′ exoribonuclease activity.
FIG. 3.
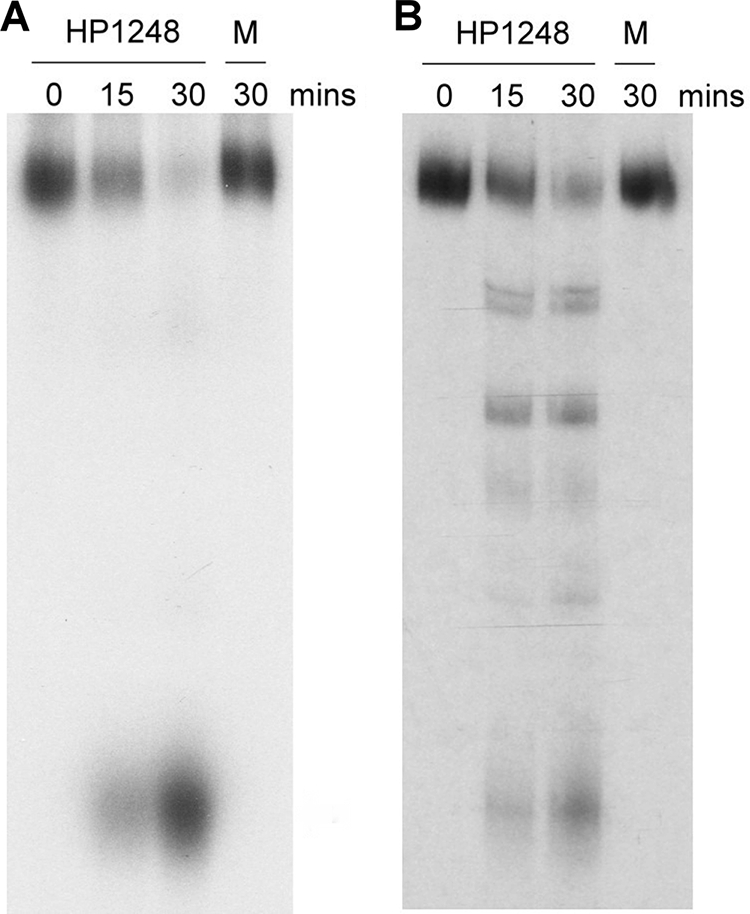
Degradation of a 5′-labeled or 3′-labeled 20-nt synthetic RNA by the HP1248 protein. (A) Degradation of 3′-labeled unmodified RNA. For the times indicated, 0.5 pmol RNA labeled at the 3′ end was incubated with 0.1 mg His-tagged HP1248. A control reaction (M) was performed with the HP1248 protein D231N mutant for 30 min. (B) Degradation of 5′-labeled unmodified RNA. For the times indicated, 0.5 pmol RNA labeled at the 5′ end was incubated with 0.1 mg His-tagged HP1248. A control reaction (M) was performed with the HP1248 protein D231N mutant for 30 min.
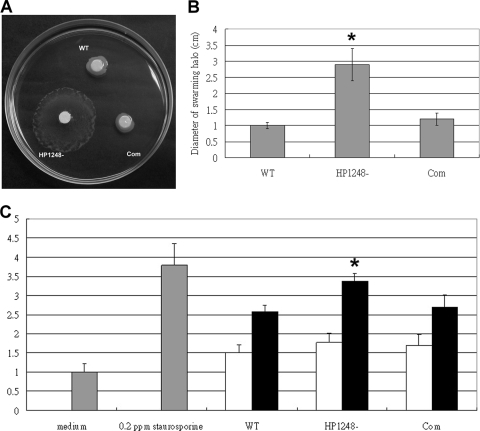
The H. pylori HP1248 deletion mutant is more motile and induces more macrophage apoptosis.
Because the motility-related genes (flaB and HP1192) were upregulated in the C1/HP1248-cat deletion mutant, we wanted to know whether the HP1248 deletion mutant could affect the motility of H. pylori. Therefore, soft agar motility tests were performed to compare the motilities of H. pylori wild-type and C1/HP1248-cat deletion mutant strains. To do this, cells were spotted onto 4-mm-diameter sterilized filter papers, and then these filter papers were put on low-concentration agar plates and incubated for 4 days at 37°C under microaerophilic conditions. The diameter of the swarming halo was measured. The results are shown in Fig. 4A and B. The areas of swarming halo of the C1/HP1248-cat deletion mutant strain were significantly increased over those of the wild-type strain, thus showing an increase of motility functions. Complementation of HP1248 restored the spreading phenotype to a level similar to that of the wild-type strain. We concluded that H. pylori lacking HP1248 is more motile.
FIG. 4.
H. pylori strain lacking HP1248 is more motile and induces more macrophage apoptosis. (A) The C1/HP1248-cat mutant strain formed a larger swarming halo than the wild-type and complementation strains. The photograph shows a Brucella broth-10% FBS soft agar plate after 4 days of incubation. WT, HP1248−, and Com indicate the wild type, C1/HP1248-cat mutant strain, and HP1248 complementation strain, respectively. (B) Quantitative analysis of the diameter of the swarming halo (cm). Values are expressed as means ± standard deviations for experiments performed in triplicate. The HP1248− mutant strain formed a larger swarming halo than the wild type (*, P < 0.01, Student's unpaired t test). (C) RAW 264.7 cells infected with H. pylori at an MOI of 50:1 (black bars) or 30:1 (white bars). RAW 264.7 cells infected with H. pylori wild-type NTUH-C1 (WT) showed an increase in apoptosis in comparison with results for uninfected controls (medium). However, the C1/HP1248-cat deletion mutant strain (HP1248−) showed an increase in apoptosis compared to results for RAW 264.7 cells infected with the wild-type strain at an MOI of 50:1 (*, P = 0.0107, Student's unpaired t test). The relative apoptosis level of HP1248− is 3.37 ± 0.21 at an MOI of 50:1 and 1.77 ± 0.25 at an MOI of 30:1. Complementation of HP1248− (Com) can decrease the ability to induce apoptosis to the WT level. Macrophage apoptosis levels were assessed by ELISA. The OD of unstimulated control cells (medium only) was assigned a value of 1.0, and the relative amounts of apoptosis in experimental groups were determined as ratios to the control level. Values are expressed as means ± standard deviations for experiments performed in triplicate. Staurosporine (0.2 ppm) was used as a positive control.
In addition, because HP1248 can also downregulate apoptosis-inducing factors (HP0175, gtt, and cagA), we wanted to know whether the HP1248 deletion mutant could affect the apoptosis-inducing ability of H. pylori. To identify the effect of HP1248 on the apoptosis of macrophages, we performed an apoptosis ELISA using the wild type, the HP1248 deletion mutant, and the complementation strain in three separate experiments. As shown in Fig. 4C, apoptosis induction by the H. pylori C1/HP1248-cat deletion mutant was efficiently increased compared to those by the wild-type and complementation strains at an MOI of 50:1. Our results were in agreement with those of a previous study (30); the level of apoptosis at an MOI of 30:1 was not statistically significant. On the other hand, we also observed that the HP1248 deletion mutant in strain 26695 is more motile and induces more macrophage apoptosis (data not shown). These findings indicate that HP1248 is involved in H. pylori inducing apoptosis of macrophages.
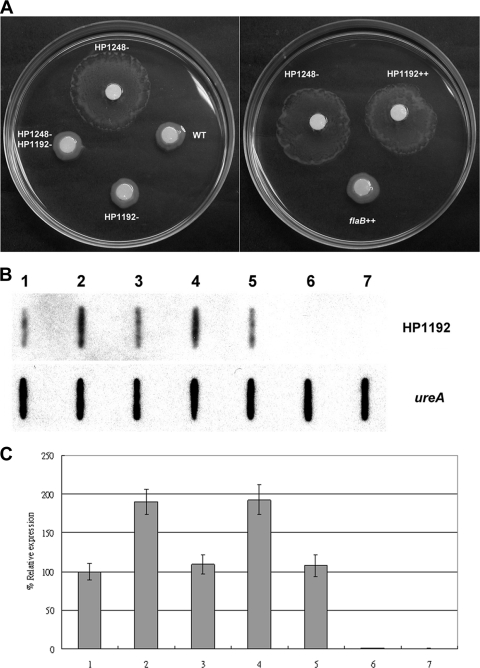
HP1192 is associated with H. pylori motility.
Because H. pylori lacking HP1248 is significantly more motile, we wanted to know whether the increased motility in the HP1248 deletion mutant was due to the upregulation of motility-related genes (HP1192 and flaB). We measured the motilities of the H. pylori wild-type, flaB overexpression, HP1192 overexpression, C1/HP1248-cat deletion mutant, C1/HP1192-km deletion mutant, and C1/HP1248-cat/HP1192-km double deletion mutant strains. Figure 5A shows that the HP1248 deletion mutant consistently produced an increased the swarm halo compared to the wild-type strain, but knockout of HP1192 in the HP1248 deletion mutant returned the motility to the level for the wild-type strain. Interestingly, the HP1192 overexpression strain produced a larger halo, the same as the HP1248 deletion mutant, but the flaB overexpression strain did not. As shown in Fig. 5B and C, the HP1248 deletion mutant and HP1192 overexpression strains, which produce larger swarm halos than the wild type, also produce a higher level of HP1192 RNA expression than the wild type. Consequently, we concluded that expression of HP1192 is associated with H. pylori motility and this might be through the regulation of HP1248.
FIG. 5.
HP1192 is required for H. pylori motility. (A) Photograph of a Brucella broth-10% FBS soft agar plate after 4 days of incubation. WT, HP1248−, HP1192−, and HP1248− HP1192− indicate the wild type, C1/HP1248-cat mutant strain, C1/HP1192-km deletion mutant strain, and HP1248/HP1192 double mutant, respectively. HP1192++ and flaB++ indicate the HP1192 overexpression strain and flaB overexpression strain, respectively. (B) Slot blot. The amount of RNA was standardized at 10 μg per slot. RNA extracted from the H. pylori WT (lane 1), HP1248 deletion mutant (lane 2), HP1248 complementation strain (lane 3), HP1192 overexpression strain (lane 4), flaB overexpression strain (lane 5), HP1192 deletion mutant (lane 6), or HP1248− HP1192− double mutant (lane 7) was assayed in the respective experiments performed with probe specific for HP1192 genes. RNA slot blot analysis with labeled probes specific for ureA was performed as an internal control. (C) Quantitative analysis of triplicate HP1192 RNA expression normalized with ureA RNA expression.
The presence of the HP1248 protein and a specific RNase recognition sequence in mRNA can reduce gene expression in H. pylori.
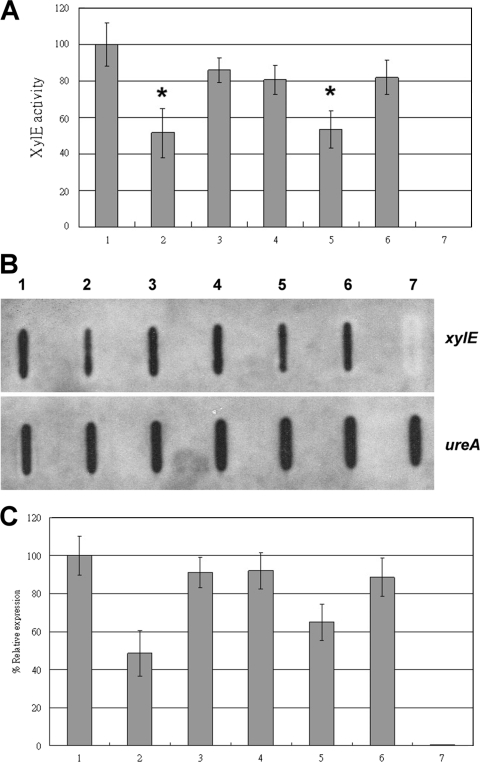
In previous studies, mRNA fragments containing a specific sequence (the REP sequence) have been shown to be substrates of RNase R in E. coli (11). To verify the mechanism through which HP1248 degrades specific mRNA, we compared the sequences of virulence-related genes which were posttranscriptionally downregulated by the HP1248 protein, including HP0175, gtt, cagA, flaB, cheY, and HP1192. We observed that HP0175, HP1192, and cagA of NTUH-C1 had similar specific RR sequences: 5′-GTGAGT(N)6-9ACTCAC-3′ in the positions +626 to +644, +46 to +66, and +3179 to +3196 relative to the transcriptional start sites of HP0175, HP1192, and cagA, respectively.
To examine the role of this specific RR sequence in the degradation of specific mRNA by HP1248 in vivo, we used the pHel3/flaA′-xylE plasmid and designed the specific RR sequences in the positions +801 to +820 relative to the transcriptional start site of the xylE reporter gene by site-directed mutagenesis. A protein domain search (http://www.ncbi.nlm.nih.gov/Structure/cdd/wrpsb.cgi) revealed that this position did not contain any functional domain of XylE. The flaA′ sequence is the predicted promoter of the flaA gene, which is an active promoter in H. pylori (7). These reporter genes with or without a specific RR sequence on the shuttle vector pHel3 were introduced into H. pylori strains NTUH-C1, C1/HP1248-cat, C1/HP1248-cat complemented with pHel3, and C1/HP1248-cat complemented by D231N with pHel3; then, the XylE activity expressed in each strain was measured in three separate experiments. As shown in Fig. 6A, the activity of XylE with the specific RR sequence was reduced markedly in NTUH-C1 compared with that without the specific RR sequence. However, the activities of XylE with and without the specific RR sequence were the same in the HP1248 deletion mutant. In addition, the activities of XylE with and without the specific RR sequence were not different in C1/HP1248-cat, showing that the specific RR sequence in the xylE gene did not affect XylE activity. The reduction of XylE activity in the presence of the specific RR sequence in the wild type was lost after deletion of HP1248. We further complemented HP1248 or D231N in the C1/HP1248-cat plasmid with the specific RR sequence. Complementation with HP1248 again reduced the activity of XylE with the specific RR sequence, whereas complementation with D231N did not. Thus, these results indicated that expression of xylE will be reduced only when the specific RR sequence of xylE and HP1248 coexist and that the exonucleolytic activity of HP1248 is required for xylE gene repression.
FIG. 6.
(A) XylE activity of H. pylori flaA′-xylE transcriptional reporter strains. Specific XylE activities were determined by using bacteria that had been grown for 48 h. XylE activity of NTUH-C1/pHel3/flaA′-xylE was assigned a value of 100, and levels of XylE activity in experimental groups were determined as a ratio to that of NTUH-C1/pHel3/flaA′-xylE. Results represent means ± standard deviations for three assays. *, P < 0.01 versus results for NTUH-C1/pHel3/flaA′-xylE. Levels of XylE activity were significantly lower for strain NTUH-C1 harboring pHel3/flaA′-xylE with the specific RR sequence (lane 2) than in strain NTUH-C1 harboring pHel3/flaA′-xylE without the specific RR sequence (lane 1); *, P < 0.001. C1/HP1248-cat harboring pHel3/flaA′-xylE without the specific RR sequence (lane 3), C1/HP1248-cat harboring pHel3/flaA′-xylE with the specific RR sequence (lane 4), complementation of C1/HP1248-cat harboring pHel3/flaA′-xylE with the specific RR sequence (lane 5), D231N complementation of C1/HP1248-cat harboring pHel3/flaA′-xylE with the specific RR sequence (lane 6), and NTUH-C1 harboring pHel3 only (lane 7) were also detected in this assay. (B) Slot blot. The amount of RNA was standardized at 10 μg per slot. RNA extracted from H. pylori (the definitions of lanes 1 to 7 are the same as those for panel A) was used in the respective experiments performed with probe specific for xylE genes. RNA slot blot analysis with labeled probes specific for ureA was performed as an internal control. (C) Quantitative analysis of triplicate xylE RNA expression normalized with ureA RNA expression.
As shown in Fig. 6B and C, the RNA expression level of xylE in the strains described above was correlated to the activity of XylE. These data suggested that the xylE gene is regulated by the degradation of xylE mRNA by the exonucleolytic activity of HP1248 and that the specific RR sequence could be the recognition site of the HP1248 protein.
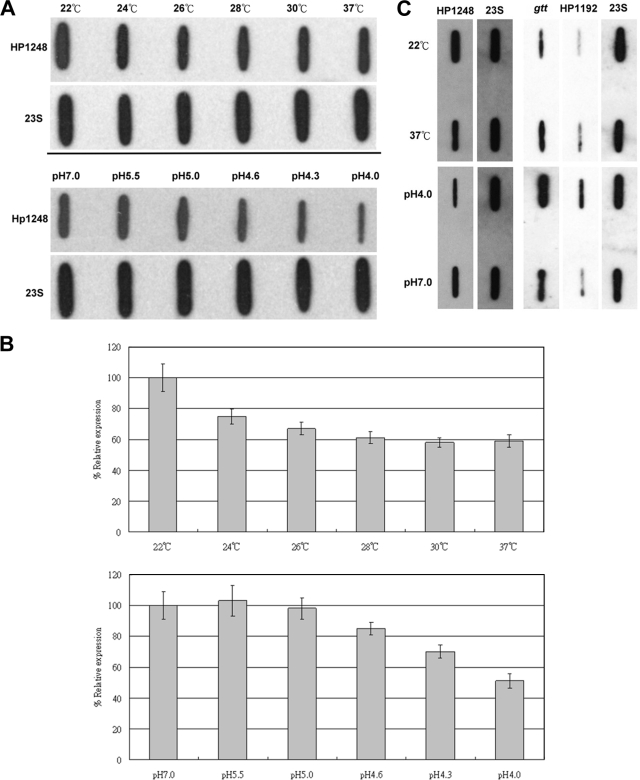
Temperature and pH can affect HP1248 expression in H. pylori.
In a recent study (8), the RNase R protein of E. coli was reported to be regulated by cold shock and RNase R of E. coli was particularly important under stress conditions. To determine whether the RNA level of HP1248 was subjected to some kind of regulation under different temperature or pH conditions, lower-temperature or lower-pH conditions were tested. Slot blot analysis revealed that the mRNA encoding HP1248 is induced upon lower-temperature or higher-pH conditions(Fig. 7A and B). Compared with results at 37°C, the HP1248 levels increased by an average of about 1.7-fold when at 22°C. Compared with results at pH 4.0, the HP1248 levels increased by an average of about twofold at pH 7.0. The data showed that only a temperature below 28°C or pH 5.0 can trigger the change in the RNA expression level of HP1248 (Fig. 7B). In addition, we also confirmed the RNA expression levels of gtt and HP1192 at different pHs and temperatures. Slot blot analysis revealed that the mRNA-encoding HP1248 target genes (gtt and HP1192) were repressed under lower-temperature or higher-pH conditions (Fig. 7C), as predicted by the changes in HP1248 levels. These results showed that RNA expression levels of HP1248 were affected by temperature and pH.
FIG. 7.
(A) Effects of temperature and pH on RNA expression levels of HP1248. Equal amounts of RNA extracted from the H. pylori NTUH-C1 strain incubated at different pHs and different temperatures for 4 h were used in the experiments performed with probe specific for HP1248. RNA slot blot analysis with a labeled probe specific for 23S RNA was performed as an internal control. (B) Quantitative analysis of triplicate HP1248 RNA expression normalized with 23S RNA expression. The expression level of HP1248 at 22°C and pH 7.0 was assigned a value of 100. (C) Effects of temperature and pH on the RNA expression levels of HP1248, HP1192, and gtt. Equal amounts of RNA extracted from the H. pylori NTUH-C1 strain incubated at different pHs and different temperatures for 4 h were used in the experiments performed with probe specific for the indicated genes. RNA slot blot analysis with a labeled probe specific for 23S RNA was performed as an internal control.
DISCUSSION
It was previously shown that vacB is associated with the expression of virulence in Shigella and enteroinvasive E. coli (12), but the mechanism was not determined. E. coli vacB has been found to encode RNase R (12). So, we suggested that HP1248 might encode RNase R and that mRNA decay from HP1248 RNase activity might be one of the regulatory mechanisms of some virulence factors in H. pylori. Therefore, we first characterized which virulence factors were downregulated by HP1248 by using transcriptional profiling of H. pylori NTUH-C1 and the C1/HP1248-cat mutant. There are 27 open reading frames that increased their expression levels significantly (>2-fold) in the HP1248 deletion mutant. This implied the fact that some genes are important or have essential functions in bacterial physiology and pathology. For example, HP1192 and flaB were found to be highly expressed in the HP1248 deletion mutant, and the flaB gene has been shown to be essential for H. pylori motility and colonization. These genes allow a bacterium to move through the viscous mucus covering the epithelial cells of the gastric mucosa. H. pylori chemotaxis modulates inflammation in mice, and the cheY mutant loses chemotaxis but retains motility (29). In addition, HP0175, gtt, and cagA were also found to be highly expressed in the HP1248 mutant, and these genes have been shown to be apoptosis-inducing factors of H. pylori. Motility and host cell apoptosis are very important to H. pylori pathogenesis. We suggested that HP1248 might play a key role in virulence factor regulation in H. pylori pathogenesis. However, we also identified 21 genes downregulated in the C1/HP1248-cat deletion mutant. We suggest that these 21 genes could be regulated indirectly by HP1248. The association of other upregulated genes and downregulated genes with HP1248 needs further investigation.
In the HP1248 deletion mutant, levels of production of the six virulence genes, HP0175, gtt, cagA, flaB, cheY, and HP1192, all increased above those in the wild type. Furthermore, the activities of the promoters of these six virulence genes in the HP1248 deletion mutant were the same as those in the wild type regardless of the presence of HP1248. Therefore, it was suggested that HP1248 might be involved in the expression of the virulence genes HP0175, gtt, cagA, flaB, cheY, and HP1192 at the posttranscriptional level.
There are no studies describing HP1248 of H. pylori as having RNase activity; however, the analogous RNase R of E. coli acts nonspecifically on poly(U) and rRNA in vitro (21). In this work, we used an HP1192 runoff transcript and 20-nt synthetic RNA as the RNA substrate. The results revealed that the HP1248 protein displayed good activity against the RNA we used. Several experiments provided convincing evidence that 3′-to-5′ exoribonuclease activity is associated with the HP1248 protein. We can detect exoribonuclease activity with the purified recombinant HP1248 protein, and the generation of mononucleotides by the HP1248 protein is completely abolished by a mutation (D231N) in the predicted active site of the enzyme.
HP1248 (RNase R) is very highly conserved throughout the bacteria. Thus, degradation of mRNA of virulence genes by a 3′-to-5′ exoribonucleolytic pathway could be a very widespread property of prokaryotes. Based upon the information presented here, we propose that HP1248 be named rnr.
In this study, motility and apoptosis-inducing ability were increased in the H. pylori HP1248 deletion mutant. The flaB gene and HP1192 were upregulated about threefold. Interestingly, the HP1192 overexpression strain was more motile than the wild-type strain but the flaB overexpression strain was not. According to the database (http://cmr.jcvi.org/cgi-bin/CMR/CmrHomePage.cgi), HP1192 is probably a motility protein-related protein, but no study has investigated the role of HP1192 in H. pylori motility. In this study, we suggested the improvement of motility conferred by HP1192 might be through the regulation of HP1248.
Recently mRNA fragments containing REP sequences have also been shown to be substrates of RNase R (11). However, the reported RNase R activity is visible only with the combined absence of PNPase, demonstrating overlapping functions of these proteins in E. coli. Another study showed that some genes without REP sequences are also regulated by RNase R in E. coli (3). In our study, the six virulence genes regulated by HP1248 did not have REP sequences. However, we found that HP0175, HP1192, and cagA of NTUH-C1 had a putative RR sequence, which was regulated by HP1248, as shown by our reporter studies. The RR sequences were predicted by software (RNAstructure v4.5) to form stem-loop secondary structures and could be the recognition sites for the HP1248 protein. Therefore, whether there were actually stem-loops responsible for the RNase recognition or other mechanisms were involved needs future study. However, gtt, flaB, and cheY did not have apparent specific RR sequences. Other explanations may be forthcoming, since the role of RNase R in mRNA decay pathways is not yet fully understood. In E. coli, some secondary structures are structural elements in mRNA that protect 3′-to-5′ exonucleolytic degradation, but RNase R can digest through extensive secondary structure provided there is a single-stranded region, such as a poly(A) tail, to which it can bind (22). HP0640 is a predicted poly(A) polymerase in H. pylori. We suggest that the poly(A) polymerase of H. pylori may also play some role in gene regulation by mRNA decay. HP0640 could provide a poly(A) tail to which the HP1248 protein could bind, and the specific RR sequence could be the signal sequence.
It has been shown that RNase R of E. coli is a cold shock protein (8). The slot blot results showed that HP1248 can be affected by pH and temperature (Fig. 7B). To H. pylori, the pH and temperature are very different between the human stomach and the outside environment. These environmental conditions may affect when H. pylori expresses the virulence factors to infect the host. On the other hand, because RNase R was also described as a gene associated with the full expression of the virulence phenotype of Shigella spp. and enteroinvasive E. coli (12), we suggest that HP1248 may be involved in some mechanisms of H. pylori pathogenesis associated with temperature and pH conditions.
Acknowledgments
This study was supported by grants from the National Science Council of Taiwan.
Footnotes
Published ahead of print on 13 February 2009.
REFERENCES
- 1.Alamuri, P., and R. J. Maier. 2004. Methionine sulphoxide reductase is an important antioxidant enzyme in the gastric pathogen Helicobacter pylori. Mol. Microbiol. 531397-1406. [DOI] [PubMed] [Google Scholar]
- 2.Amblar, M., and C. M. Arraiano. 2005. A single mutation in Escherichia coli ribonuclease II inactivates the enzyme without affecting RNA binding. FEBS J. 272363-374. [DOI] [PubMed] [Google Scholar]
- 3.Andrade, J. M., F. Cairrao, and C. M. Arraiano. 2006. RNase R affects gene expression in stationary phase: regulation of ompA. Mol. Microbiol. 60219-228. [DOI] [PubMed] [Google Scholar]
- 4.Ang, S., C. Z. Lee, K. Peck, M. Sindici, U. Matrubutham, M. A. Gleeson, and J. T. Wang. 2001. Acid-induced gene expression in Helicobacter pylori: study in genomic scale by microarray. Infect. Immun. 691679-1686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Basak, C., S. K. Pathak, A. Bhattacharyya, S. Pathak, J. Basu, and M. Kundu. 2005. The secreted peptidyl prolyl cis,trans-isomerase HP0175 of Helicobacter pylori induces apoptosis of gastric epithelial cells in a TLR4- and apoptosis signal-regulating kinase 1-dependent manner. J. Immunol. 1745672-5680. [DOI] [PubMed] [Google Scholar]
- 6.Blaser, M. J. 1992. Hypotheses on the pathogenesis and natural history of Helicobacter pylori-induced inflammation. Gastroenterology 102720-727. [DOI] [PubMed] [Google Scholar]
- 7.Brahmachary, P., M. G. Dashti, J. W. Olson, and T. R. Hoover. 2004. Helicobacter pylori FlgR is an enhancer-independent activator of sigma54-RNA polymerase holoenzyme. J. Bacteriol. 1864535-4542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cairrao, F., A. Cruz, H. Mori, and C. M. Arraiano. 2003. Cold shock induction of RNase R and its role in the maturation of the quality control mediator SsrA/tmRNA. Mol. Microbiol. 501349-1360. [DOI] [PubMed] [Google Scholar]
- 9.Censini, S., C. Lange, Z. Xiang, J. E. Crabtree, P. Ghiara, M. Borodovsky, R. Rappuoli, and A. Covacci. 1996. cag, a pathogenicity island of Helicobacter pylori, encodes type I-specific and disease-associated virulence factors. Proc. Natl. Acad. Sci. USA 9314648-14653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chang, K. C., S. W. Ho, J. C. Yang, and J. T. Wang. 1997. Isolation of a genetic locus associated with metronidazole resistance in Helicobacter pylori. Biochem. Biophys. Res. Commun. 236785-788. [DOI] [PubMed] [Google Scholar]
- 11.Cheng, Z. F., and M. P. Deutscher. 2005. An important role for RNase R in mRNA decay. Mol. Cell 17313-318. [DOI] [PubMed] [Google Scholar]
- 12.Cheng, Z. F., Y. Zuo, Z. Li, K. E. Rudd, and M. P. Deutscher. 1998. The vacB gene required for virulence in Shigella flexneri and Escherichia coli encodes the exoribonuclease RNase R. J. Biol. Chem. 27314077-14080. [DOI] [PubMed] [Google Scholar]
- 13.Chevalier, C., J. M. Thiberge, R. L. Ferrero, and A. Labigne. 1999. Essential role of Helicobacter pylori gamma-glutamyltranspeptidase for the colonization of the gastric mucosa of mice. Mol. Microbiol. 311359-1372. [DOI] [PubMed] [Google Scholar]
- 14.Covacci, A., J. L. Telford, G. Del Giudice, J. Parsonnet, and R. Rappuoli. 1999. Helicobacter pylori virulence and genetic geography. Science 2841328-1333. [DOI] [PubMed] [Google Scholar]
- 15.Eaton, K. A., D. R. Morgan, and S. Krakowka. 1992. Motility as a factor in the colonisation of gnotobiotic piglets by Helicobacter pylori. J. Med. Microbiol. 37123-127. [DOI] [PubMed] [Google Scholar]
- 16.Fan, X. G., D. Kelleher, X. J. Fan, H. X. Xia, and P. W. Keeling. 1996. Helicobacter pylori increases proliferation of gastric epithelial cells. Gut 3819-22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gobert, A. P., Y. Cheng, J. Y. Wang, J. L. Boucher, R. K. Iyer, S. D. Cederbaum, R. A. Casero, Jr., J. C. Newton, and K. T. Wilson. 2002. Helicobacter pylori induces macrophage apoptosis by activation of arginase II. J. Immunol. 1684692-4700. [DOI] [PubMed] [Google Scholar]
- 18.Heuermann, D., and R. Haas. 1998. A stable shuttle vector system for efficient genetic complementation of Helicobacter pylori strains by transformation and conjugation. Mol. Gen. Genet. 257519-528. [DOI] [PubMed] [Google Scholar]
- 19.Hsieh, P. F., J. C. Yang, J. T. Lin, and J. T. Wang. 1998. Molecular mechanisms of clarithromycin resistance in Helicobacter pylori. J. Formos. Med. Assoc. 97445-452. [PubMed] [Google Scholar]
- 20.Karzai, A. W., and R. T. Sauer. 2001. Protein factors associated with the SsrA. SmpB tagging and ribosome rescue complex. Proc. Natl. Acad. Sci. USA 983040-3044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kasai, T., R. S. Gupta, and D. Schlessinger. 1977. Exoribonucleases in wild type Escherichia coli and RNase II-deficient mutants. J. Biol. Chem. 2528950-8956. [PubMed] [Google Scholar]
- 22.Khemici, V., and A. J. Carpousis. 2004. The RNA degradosome and poly(A) polymerase of Escherichia coli are required in vivo for the degradation of small mRNA decay intermediates containing REP-stabilizers. Mol. Microbiol. 51777-790. [DOI] [PubMed] [Google Scholar]
- 23.Kim, K. M., S. G. Lee, M. G. Park, J. Y. Song, H. L. Kang, W. K. Lee, M. J. Cho, K. H. Rhee, H. S. Youn, and S. C. Baik. 2007. Gamma-glutamyltranspeptidase of Helicobacter pylori induces mitochondria-mediated apoptosis in AGS cells. Biochem. Biophys. Res. Commun. 355562-567. [DOI] [PubMed] [Google Scholar]
- 24.Kostrzynska, M., J. D. Betts, J. W. Austin, and T. J. Trust. 1991. Identification, characterization, and spatial localization of two flagellin species in Helicobacter pylori flagella. J. Bacteriol. 173937-946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Le'Negrate, G., V. Ricci, V. Hofman, B. Mograbi, P. Hofman, and B. Rossi. 2001. Epithelial intestinal cell apoptosis induced by Helicobacter pylori depends on expression of the cag pathogenicity island phenotype. Infect. Immun. 695001-5009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Li, Z., S. Reimers, S. Pandit, and M. P. Deutscher. 2002. RNA quality control: degradation of defective transfer RNA. EMBO J. 211132-1138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Marshall, B. J., and J. R. Warren. 1984. Unidentified curved bacilli in the stomach of patients with gastritis and peptic ulceration. Lancet i1311-1315. [DOI] [PubMed] [Google Scholar]
- 28.Mathy, N., L. Benard, O. Pellegrini, R. Daou, T. Wen, and C. Condon. 2007. 5′-to-3′ exoribonuclease activity in bacteria: role of RNase J1 in rRNA maturation and 5′ stability of mRNA. Cell 129681-692. [DOI] [PubMed] [Google Scholar]
- 29.McGee, D. J., M. L. Langford, E. L. Watson, J. E. Carter, Y. T. Chen, and K. M. Ottemann. 2005. Colonization and inflammation deficiencies in Mongolian gerbils infected by Helicobacter pylori chemotaxis mutants. Infect. Immun. 731820-1827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Menaker, R. J., P. J. Ceponis, and N. L. Jones. 2004. Helicobacter pylori induces apoptosis of macrophages in association with alterations in the mitochondrial pathway. Infect. Immun. 722889-2898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Moss, S. F. 1998. Helicobacter pylori and apoptosis. Yale J. Biol. Med. 7153-61. [PMC free article] [PubMed] [Google Scholar]
- 32.Nomura, A., G. N. Stemmermann, P. H. Chyou, I. Kato, G. I. Perez-Perez, and M. J. Blaser. 1991. Helicobacter pylori infection and gastric carcinoma among Japanese Americans in Hawaii. N. Engl. J. Med. 3251132-1136. [DOI] [PubMed] [Google Scholar]
- 33.Ottemann, K. M., and A. C. Lowenthal. 2002. Helicobacter pylori uses motility for initial colonization and to attain robust infection. Infect. Immun. 701984-1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Oussenko, I. A., R. Sanchez, and D. H. Bechhofer. 2002. Bacillus subtilis YhaM, a member of a new family of 3′-to-5′ exonucleases in gram-positive bacteria. J. Bacteriol. 1846250-6259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Parsonnet, J., G. D. Friedman, D. P. Vandersteen, Y. Chang, J. H. Vogelman, N. Orentreich, and R. K. Sibley. 1991. Helicobacter pylori infection and the risk of gastric carcinoma. N. Engl. J. Med. 3251127-1131. [DOI] [PubMed] [Google Scholar]
- 36.Rader, B. A., S. R. Campagna, M. F. Semmelhack, B. L. Bassler, and K. Guillemin. 2007. The quorum-sensing molecule autoinducer 2 regulates motility and flagellar morphogenesis in Helicobacter pylori. J. Bacteriol. 1896109-6117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Schmitz, A., C. Josenhans, and S. Suerbaum. 1997. Cloning and characterization of the Helicobacter pylori flbA gene, which codes for a membrane protein involved in coordinated expression of flagellar genes. J. Bacteriol. 179987-997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Suerbaum, S., C. Josenhans, and A. Labigne. 1993. Cloning and genetic characterization of the Helicobacter pylori and Helicobacter mustelae flaB flagellin genes and construction of H. pylori flaA- and flaB-negative mutants by electroporation-mediated allelic exchange. J. Bacteriol. 1753278-3288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Tiwari, S., U. Ghoshal, U. C. Ghoshal, S. Dhingra, R. Pandey, M. Singh, A. Ayyagari, and S. Naik. 2005. Helicobacter pylori-induced apoptosis in pathogenesis of gastric carcinoma. Indian J. Gastroenterol. 24193-196. [PubMed] [Google Scholar]
- 40.Tobe, T., C. Sasakawa, N. Okada, Y. Honma, and M. Yoshikawa. 1992. vacB, a novel chromosomal gene required for expression of virulence genes on the large plasmid of Shigella flexneri. J. Bacteriol. 1746359-6367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Williams, S. M., Y. T. Chen, T. Andermann, J. E. Carter, D. J. McGee, and K. M. Ottemann. 2007. Helicobacter pylori chemotaxis modulates inflammation and gastric-epithelium interactions in infected mice. Infect. Immun. 753747-3757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wotherspoon, A. C., C. Doglioni, T. C. Diss, L. Pan, A. Moschini, M. de Boni, and P. G. Isaacson. 1993. Regression of primary low-grade B-cell gastric lymphoma of mucosa-associated lymphoid tissue type after eradication of Helicobacter pylori. Lancet 342575-577. [DOI] [PubMed] [Google Scholar]
- 43.Xu, Q., and M. J. Blaser. 2001. Promoters of the CATG-specific methyltransferase gene hpyIM differ between iceA1 and iceA2 Helicobacter pylori strains. J. Bacteriol. 1833875-3884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Yeh, Y. C., T. L. Lin, K. C. Chang, and J. T. Wang. 2003. Characterization of a ComE3 homologue essential for DNA transformation in Helicobacter pylori. Infect. Immun. 715427-5431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Zhu, Y., C. Wang, J. Huang, Z. Ge, Q. Dong, X. Zhong, Y. Su, and S. Zheng. 2007. The Helicobacter pylori virulence factor CagA promotes Erk1/2-mediated Bad phosphorylation in lymphocytes: a mechanism of CagA-inhibited lymphocyte apoptosis. Cell Microbiol. 9952-961. [DOI] [PubMed] [Google Scholar]
- 46.Zukowski, M. M., D. F. Gaffney, D. Speck, M. Kauffmann, A. Findeli, A. Wisecup, and J. P. Lecocq. 1983. Chromogenic identification of genetic regulatory signals in Bacillus subtilis based on expression of a cloned Pseudomonas gene. Proc. Natl. Acad. Sci. USA 801101-1105. [DOI] [PMC free article] [PubMed] [Google Scholar]