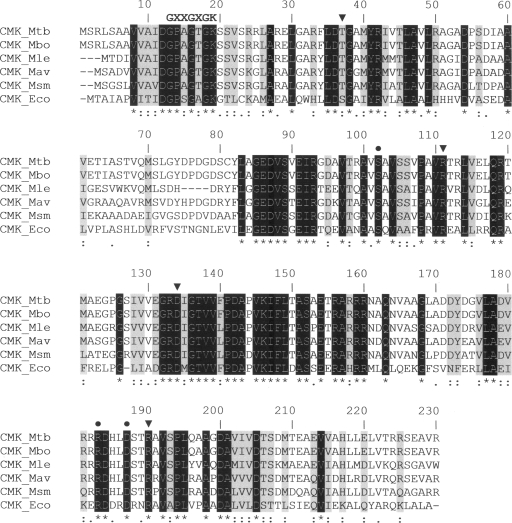
FIG. 2.
Multiple sequence alignment of mycobacterial CMKs with the known three-dimensional structure of E. coli CMK (CMK_Eco). The putative CMK sequences of M. tuberculosis (Mtb), M. bovis (Mbo), M. leprae (Mle), M. avium (Mav), and M. smegmatis (Msm) were aligned with CMK_Eco by using the default settings of the CLUSTALW program (18). Identical conserved residues are shown in white on a black background and are also indicated by asterisks below the alignment. Strongly similar and weakly similar residues are identified by colons and periods, respectively. Strongly similar residues are also shaded in gray. Triangles and circles above the alignment indicate residues that interact with the pyrimidine ring of CMP (Ser36, Arg110, Asp132, and Arg188 in CMK_Eco) and those that are key in pentose recognition (Ser101, Arg181, and Asp185 in CMK_Eco), respectively, as determined by site-directed mutagenesis and crystallographic studies (1, 13). The conserved fingerprint sequence GXXGXGK (where X stands for any amino acid) of NMP kinases, which forms the phosphate-binding loop (P loop), is indicated at the N-terminal portion of CMKs (6).