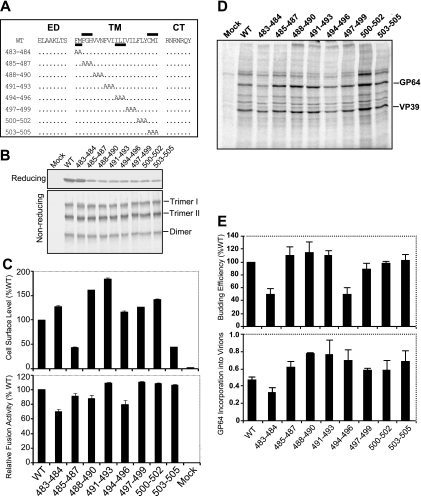
FIG. 6.
Analysis of alanine scanning mutations in the GP64 TM domain. (A) Schematic representation of the positions of GP64 alanine scanning mutations. Sets of two or three amino acids in the TM domain were replaced with alanines as indicated (dots represent positions with no change). The name of each alanine scanning GP64 construct is indicated on the left (numbers represent positions of modified amino acids in the AcMNPV GP64 sequence). The two regions important for cell surface expression of GP64 are indicated by bars above the WT GP64 TM domain sequence, and two regions important for virus budding are similarly indicated by bars below the sequence. ED, ectodomain; CT, cytoplasmic tail. (B) Expression and trimerization of GP64 constructs with alanine scanning mutations in the TM domain. Sf9 cells were infected with viruses expressing WT GP64 or the alanine scanning constructs of GP64 shown in panel A. Expression and trimerization of GP64 were examined by Western blot analysis of cell extracts on reducing SDS-PAGE (top) or nonreducing SDS-PAGE (bottom) gel. The positions of GP64 and oligomeric forms (Trimer I and II and Dimer) are indicated at the right. (C) Relative cell surface levels and fusion activities of GP64 constructs. Relative cell surface levels of GP64 constructs were measured by cELISA of infected Sf9 cells, using mAb AcV5. Relative fusion activity of each modified GP64 construct was evaluated by measuring the efficiency of syncytium formation in infected Sf9 cells. For each sample, five fields were examined. The means and standard deviations of the results of triplicate infections are illustrated. (D) Analysis of budding efficiency and incorporation of modified GP64 constructs into budded virions. Viruses expressing modified GP64 constructs were pulse labeled with [35S]methionine, and BV purified and processed as described in Materials and Methods. The positions of GP64 and VP39 proteins are indicated at the right. Each lane represents virus purified from an equivalent volume of the cell culture supernatant. (E) Relative budding efficiencies and incorporation of GP64 into budded virions. Relative virion budding efficiency was calculated by comparison of measurements of labeled major capsid protein (VP39) in each virus. All data were normalized to the results for a virus construct expressing WT GP64 (100%). The efficiency of GP64 incorporation into virions was calculated for each virus expressing a modified GP64 protein with an alanine scanning mutation. Molar ratios of GP64:VP39 were calculated for virions of each virus based on 15 methionine residues (mutant 503-505) and 16 methionine residues (mature WT GP64 and all other mutants) or 9 methionine residues (VP39). The means and standard deviations determined from triplicate experiments are shown.