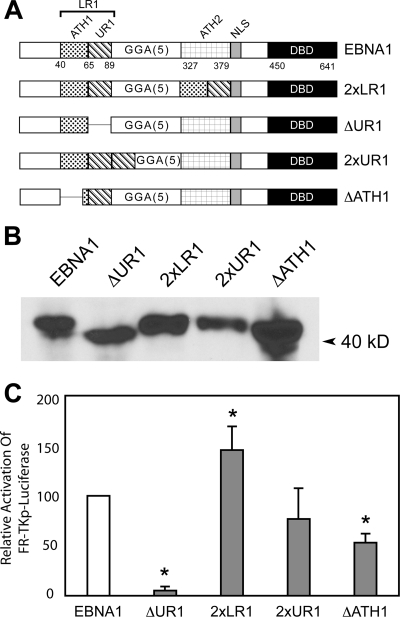
FIG. 4.
Both the UR1 and ATH1 domains within LR1 contribute to transactivation of an episomal FR-TKp-luciferase reporter in C33a cells. (A) A schematic representation of wild-type EBNA1, the previously described 2×LR1, ΔUR1 derivatives, and the 2×UR1 and ΔATH1 derivatives used in this study. 2×LR1 replaces aa 327 to 377 of EBNA1 with a second copy of aa 40 to 89. In ΔUR1 aa 71 to 88 are deleted. 2×UR1 contains a second copy of aa 65 to 89 inserted after UR1 while the GR repeat from aa 40 to 54 is deleted in ΔATH1. (B) Expression of wild-type EBNA1, ΔUR1, 2×LR1, 2×UR1, and ΔATH1 in transfected C33a cells. Proteins from 5 × 105 transfected cells were visualized as described in the Materials and Methods section, and the migration of a prestained marker is indicated by the arrowhead. (C) C33a cells were transfected with an expression plasmid for EBNA1, ΔUR1, 2×LR1, 2×UR1, or ΔATH1 proteins along with an FR-TKp-luciferase reporter and assayed for luciferase activity 48 h posttransfection. At this time, EBNA1 transactivates FR-TKp-luciferase approximately 80-fold over the DBD alone, and this value was set to 100%. In three independent transfections 2×LR1 was observed to transactivate the reporter at greater levels than wild-type EBNA1 while ΔUR1 was impaired in transactivation. Transactivation by 2×UR1 could not be distinguished from wild-type EBNA1. ΔATH1 transactivated the reporter approximately 50% as well as wild-type EBNA1. Transactivation that differs significantly from that observed with wild-type EBNA1 (P < 0.05) is indicated by the asterisk.