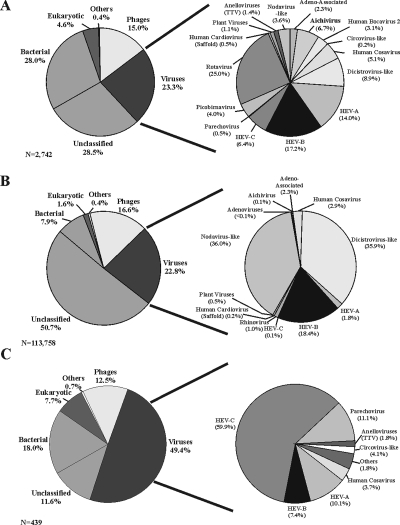
FIG. 1.
Sequence classification and distribution. (Left panels) Sequences with E values of ≤10−3 were classified as either eukaryotic, phage, bacterial, or viral based on the best tBLASTx score. Unclassified sequences are those which had E values of >10−3 by tBLASTx, BLASTn, and BLASTx analyses. Sequences which did not fall into set categories were designated “other” and included fungal and plasmid vector sequences. (Right panels) The subsets of sequences classified as viral were broken down further by family, genus, and species. Classification results for Sanger sequencing clones derived from 35 AFP patients (A), sequences generated by 454 pyrosequencing from a subset of 10 patients (B), and Sanger sequencing clones obtained from six healthy contacts (C) are shown. Others in the viral pie chart of healthy contacts consist of rhinovirus and Aichi virus.