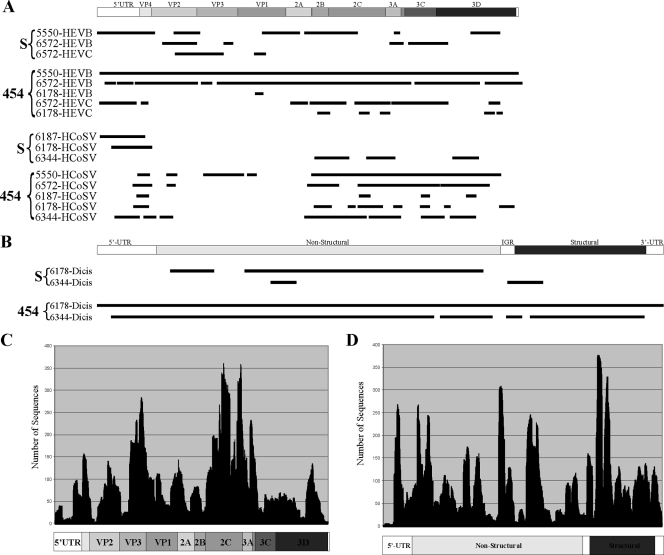
FIG. 2.
Viral genome coverage and depth of sequencing. (A) Lines below the graphical depiction of a generic picornavirus genome represent the locations of HEV-B, HEV-C, or human cosavirus (HCoSV) singlets or contigs acquired from different AFP patients by either Sanger sequencing (S) or 454 pyrosequencing (454). 5′-UTR, 5′ untranslated region; IGR, intergenomic region. (B) Lines below the graphical depiction of a generic dicistrovirus genome represent the locations of dicistrovirus (Dicis) singlets or contigs acquired from samples from two AFP patients by either Sanger sequencing or 454 pyrosequencing. (C and D) The depth of sequencing for each nucleotide position is shown for 454 pyrosequenced HEV-B from patient 5550 (C) and dicistrovirus from patient 6178 (D).