Figure 4.
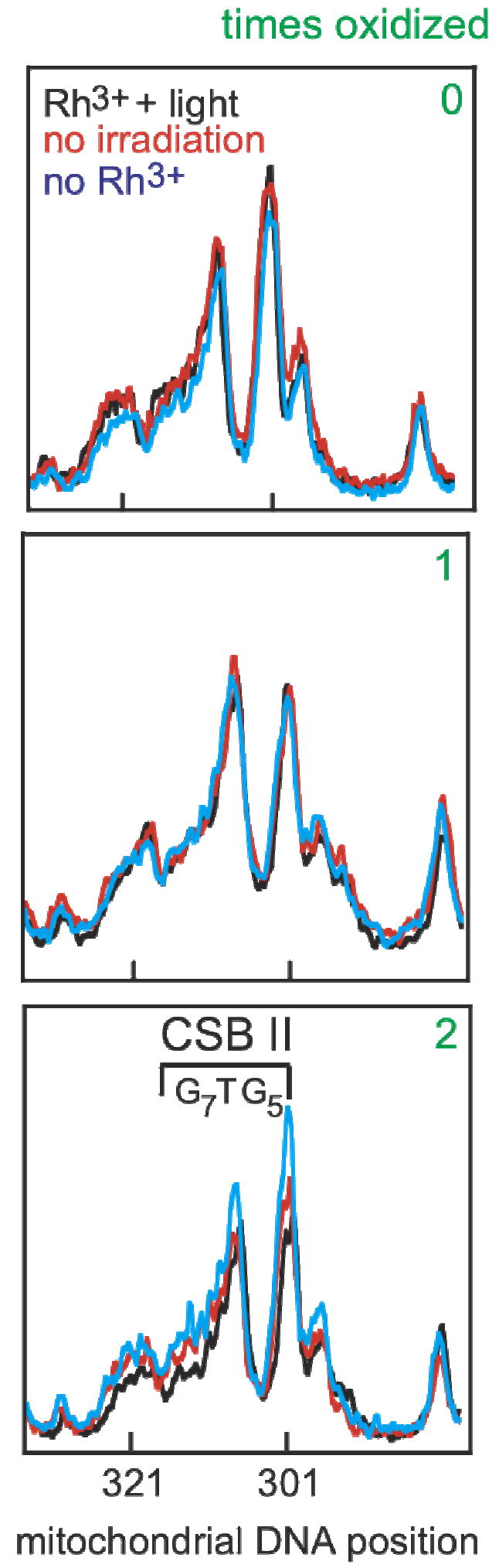
Guanine sequencing of conserved sequence block II over several cycles of oxidation and regrowth. Cellular DNA was extracted, the mitochondrial control region was amplified, and sequencing was accomplished using a 32P-labeled primer and dC terminator nucleotide. The results shown are line plots of the PAGE gels from nucleotides 284-330. Conserved sequence block II (303-315, 3’-G7AG5, marked as CSB II) is a region comprised of many guanosine residues and is prone to oxidation. A reference dG, that is not within conserved sequence block II, at position 285 is on the right of each plot. At first, cells are genetically identical, and sequencing lanes overlap (0). Cells were treated with [Rh(phi)2bpy]3+ only (no irradiation, red), only irradiated with ~365 nm light (no Rh, blue), or given Rh and irradiated (Rh+light, black) and allowed to grow. Little, if any, change in the dG content occurs in conserved sequence block II in all three cases after one cycle (labeled as 1). The same cells were treated again (labeled as 2). After two cycles, cells treated with [Rh(phi)2bpy]3+ only (red) and only irradiated (blue) have very similar dG content in conserved sequence block II. In contrast, incubation of cells with [Rh(phi)2bpy]3+ and irradiation (black) shows a decrease in the dG content in conserved sequence block II. The lack of change at position 285 with Rh and light indicates the loss of dG content is specific to conserved sequence block II.
