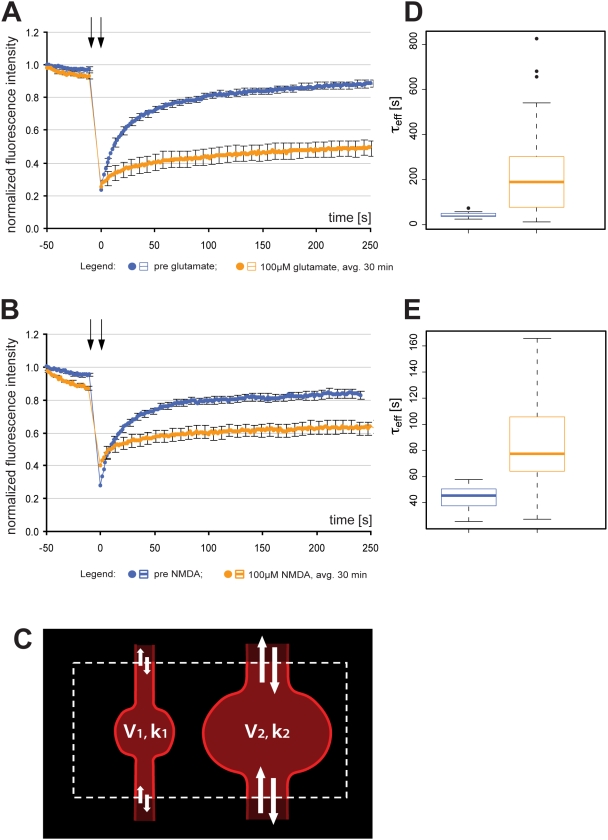
Figure 2. Analysis of ER fission by FRAP.
(A and B) Normalized average FRAP signal over time in untreated neurons (blue) and the same neurons after (A) 100 µM glutamate, or (B) 100 µM NMDA (orange). Photobleaching was performed between the arrows. Time = 0 was set to when photobleaching ends and fluorescence starts to recover. Error bars are standard error of the mean (SEM). n = 19 for glutamate; n = 17 for NMDA. (C) The ER within the ROI (dashed line) was modeled as consisting of two volumes: V1 and V2. The RedER molecules move within these volumes with rate constants k1 and k2 respectively. (D and E) Box plot of τeff values of the same neurons shown in A and B. Untreated neurons are blue and the same neurons after (D) 100 µM glutamate or (E) NMDA are orange. Note the difference in scale between D and E. The line in the box is the median and the box represents the 25–75 percentiles. Whiskers extend to the extreme values as long as they are within a range of 1.5× box length. Values outside this range are plotted as outliers. avg.: average.