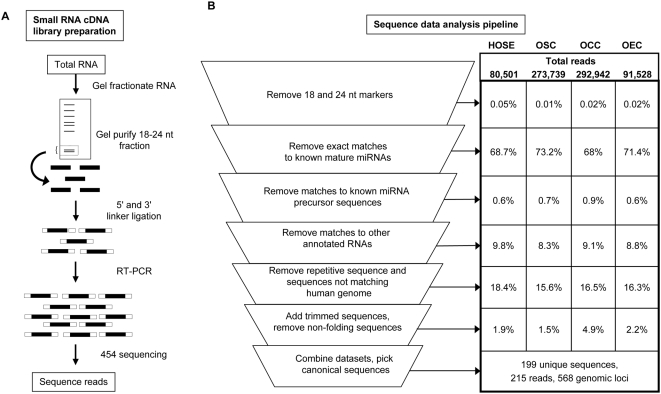
Figure 1. Small RNA cDNA library generation and data analysis pipeline.
A. Small RNAs were isolated from normal primary HOSE cultures and from serous (OSC), clear cell (OCC) and endometrioid (OEC) ovarian cancer tissues. Following 5′ and 3′ linker ligation, RT-PCR was performed to generate four independent cDNA libraries of small RNAs that were then used as templates for massively parallel pyrosequencing (454 sequencing). B. Description of the steps in the data analysis pipeline. The initial step of the data analysis was removal of sequences corresponding to 18 nt and 24 nt RNA markers that had been spiked into the total RNA prior to gel electrophoresis. Percentage of total reads from HOSE, OSC, OCC and OEC datasets classified into the designated categories and filtered out at each step are listed in the table on the right. At the bottom of the table, the number of “unique sequences” represents the non-redundant sequences derived after collapsing multiple reads of identical sequence. Some canonical sequences map to more than one locus in the genome. At completion of the analysis, 199 sequences totaling 215 reads and mapping to 568 loci met our criteria for canonical hairpin-derived sequences from the combined datasets.