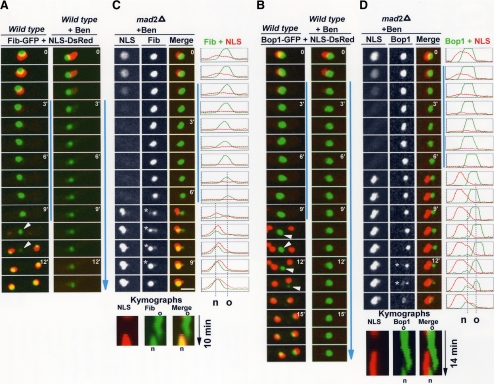
Figure 5.
Nucleolar segregation and the spindle assembly checkpoint. (A) Time-lapse images of An-Fib-GFP and NLS-DsRed during mitosis without or with addition of the microtubule poison benomyl. Mitotic entry with depolymerized microtubules activates the SAC, which maintains the mitotic dispersal of NLS-DsRed. (C) Location of An-Fib-GFP and NLS-DsRed during a spindle-independent mitosis (SIM) in a SAC-deficient strain (mad2 deletion) after addition of benomyl (Video5.mov). Line profiles of pixel intensities along a cross section and kymographs are also shown. Nuclei enter and exit mitosis as indicated by the dispersal and return of NLS-DsRed, but nuclear segregation cannot occur because of lack of spindle formation (Figure 6B). An-Fib-GFP disassembles from the old nucleolus and reassembles at a different site forming a new nucleolus (o and n in the kymographs and pixel profiles). (B and D) As for A and C, but following An-Bop1-GFP. Spindle-independent formation of a new nucleolus occurs as in C, but An-Bop1 accumulation at this site occurs significantly after NLS-DsRed reimport (see text; Video6.mov). Time points in which both the old and new nucleolus are resolved are indicated by an asterisk. Vertical blue lines indicate the period of time that cells are in mitosis. Bars, ∼5 μm.