Abstract
The putative DNA encapsidation genes encoded by open reading frames (ORFs) 25, 26, 30, 34, 43, 45/42 and 54 were cloned from Varicella-zoster virus (VZV) strain Ellen. Sequencing revealed that the Ellen ORFs were highly conserved at the amino acid level when compared to those of nineteen previously published VZV isolates. Additionally, RT-PCR provided the first evidence that ORF45/42 was expressed as a spliced transcript in VZV-infected cells. All seven ORFs were expressed in vitro and full length products were identified using a C-terminal V5 epitope tag. The in vitro products of the putative VZV terminase subunits encoded by ORFs 30 and 45/42 proved useful in protein-protein interaction assays. Previous studies have reported the formation of a heterodimeric terminase complex involved in DNA encapsidation for both herpes simplex virus-type 1 (HSV-1) and human cytomegalovirus (HCMV). Here we report that the C-terminal portion of exon II of ORF45/42 (ORF42-C269) interacted in GST-pull down experiments with in vitro synthesized ORF30 and ORF45/42. The interactions were maintained in the presence of anionic detergents and in buffers of increasing ionic strength. Cells transiently transfected with epitope tagged ORF45/42 or ORF30 showed primarily cytoplasmic staining. In contrast, an antiserum directed to the N-terminal portion of ORF45 showed nearly exclusive nuclear localization of the ORF45/42 gene product in infected cells. An ORF30 specific antiserum detected an 87 kDa protein in both the cytoplasmic and nuclear fractions of VZV infected cells. The results were consistent with the localization and function of herpesviral terminase subunits. This is the first study aimed at the identification and characterization of the VZV DNA encapsidation gene products.
Keywords: Varicella-zoster virus, DNA encapsidation, terminase, ORF30, ORF45/42
Introduction
The ongoing clinical need to prevent or treat infections caused by members of the family Herpesviridae merits continued investigation of novel antiviral agents (Biron, 2006; Bogner, 2002; Casper, 2006; Casper and Wald, 2007; De Clercq, 2001; Griffiths, 2006; Luck et al., 2006; Schang, 2006; Visalli, 2004; Visalli and van Zeijl, 2003). Proteins that play a role in the DNA encapsidation process have become promising novel targets (van Zeijl et al., 2000; Visalli et al., 2003; Visalli and van Zeijl, 2003). Studies on herpes simplex virus-type 1 (HSV-1) have contributed greatly to our understanding of the proteins involved in encapsidation. A total of seven genes have been shown to be essential in the HSV-1 DNA encapsidation process: UL's 6, 15, 17, 25, 28, 32 and 33 (Beard et al., 2002; Chang et al., 1996; Goshima et al., 2000; Koslowski et al., 1999; Lamberti and Weller, 1996; Lamberti and Weller, 1998; McNab et al., 1998; Newcomb et al., 2001a; Ogasawara et al., 2001; Patel and MacLean, 1995; Patel et al., 1996; Reynolds et al., 2000; Salmon and Baines, 1998; Sheaffer et al., 2001; Taus and Baines, 1998; Yu and Weller, 1998b). These genes are conserved throughout the herpesvirus family (Alba et al., 2001) and when any of the seven were deleted from the HSV-1 genome, empty capsids accumulated in the nucleus. There is mounting evidence that at least two of the seven encapsidation proteins form an essential terminase complex that likely functions as both an endonuclease and a DNA translocase during DNA cleavage and packaging (Bogner, 2002; Hwang and Bogner, 2002; Scheffczik et al., 2002; Scholz et al., 2003). During viral replication, progeny DNA genomes are synthesized in the nucleus as long, branched head-to-tail concatemers (Severini et al., 1996; Zhang et al., 1994). The capsid proteins are synthesized in the cytoplasm, transported to the nucleus, and assembled into procapsids (Brown, 2002). The tight packing of one unit length of herpesviral genomic DNA into preformed procapsids is likely mediated by multi-protein complexes that include the viral terminase subunits.
The human cytomegalovirus (HCMV) terminase subunits pUL56 and pUL89, encoded by the UL56 and UL89 genes, have been extensively studied. Both gene products formed toroidal structures, possessed DNA binding and nuclease activities, and specifically recognized genomic packaging (pac) sequences (Bogner et al., 1998; Scheffczik et al., 2002). The subunit pUL56 was shown to have ATPase activity that was enhanced by the addition of the pUL89 subunit (Hwang and Bogner, 2002; Scholz et al., 2003). These subunits interacted in vitro (Hwang and Bogner, 2002; Thoma et al., 2006) and could be co-immunoprecipitated from HCMV infected cell extracts (Hwang and Bogner, 2002). Additionally, pUL56 formed a complex with the HCMV portal protein, pUL104 (Dittmer et al., 2005). This latter interaction was abolished in the presence of the benzimidazole-D-ribonucleoside inhibitors BDCRB and CL4RB (Dittmer et al., 2005). It was postulated that disruption of the pUL56 / pUL104 complex was responsible for the anti-HCMV activity exhibited by these compounds. Little work has been done on the remaining HCMV encapsidation gene products.
Based on evidence for their homologous counterparts in HCMV, the UL15 and UL28 genes encode the putative HSV-1 terminase subunits. The HSV-1 UL15 and UL28 gene products were shown to interact with each other (Abbotts et al., 2000; Koslowski et al., 1999) (Jacobson et al., 2006). The UL15 / UL28 complex is presumed to direct the cleavage and packaging of genomic DNA, but neither subunit has been formally shown to exhibit ATPase activity. Detailed studies have shown that the UL6 gene encodes a portal protein found at one vertex of each preassembled capsid and that the portal is intimately involved in “guiding” viral DNA into the pre-capsid (Newcomb et al., 2001b). The HSV-1 portal protein was shown to interact with both the UL15 and UL28 proteins (White et al., 2003) and a series of thiourea compounds were shown to prevent both the UL6 and UL15 proteins from associating with capsids (Newcomb and Brown, 2002). Additionally, replicating DNA, localized to nuclear replication compartments, is probably associated with a complex containing the UL28, UL15 and UL33 gene products (Beard et al., 2002; Wills et al., 2006; Yang and Baines, 2006). The evidence thus far suggests that multiple proteins act in concert during various steps of the encapsidation process.
Very little is known about the encapsidation process in Varicella-zoster virus (VZV). The interactions previously described between the heterodimeric terminase subunits for both HSV-1 and HCMV provided a basis for our studies seeking to define protein-protein interactions important in the VZV DNA encapsidation process. This report is the first to describe the cloning, sequencing, and in vitro expression of the seven ORFs encoding the putative VZV encapsidation proteins. Consistent with studies reported previously for both HSV-1 and HCMV, the VZV terminase subunit homologs, encoded by ORF30 and ORF45/42, were shown to interact and form stable complexes in vitro and both were shown to localize in the nucleus of infected cells.
Materials and Methods
Cells and virus
Monolayer cultures of human foreskin fibroblast (HFF), human lung fibroblast (IMR-90), African green monkey kidney (Vero), or human melanoma (MeWo) cells were used for propagation of VZV strain Ellen (ATCC VR-1367). Vero, IMR-90, and HFF cells were grown in Dulbecco's Modified Eagles Medium (DME) supplemented with 10% fetal calf serum, 2 mM L-glutamine, 1.5 g/L sodium bicarbonate, 100 units/ml of penicillin, 100 mg/ml streptomycin sulfate, and 50 µg/ml ciprofloxacin. MeWo cells were grown in Minimal Essential Medium (MEM) supplemented with 8% fetal calf serum, 2 mM L-glutamine, 0.1 mM non-essential amino acids, 100 units/ml of penicillin, and 100 mg/ml streptomycin sulfate. Cells infected with VZV were incubated in either DME or MEM containing 3% serum. Infections were performed with VZV-infected cell stocks applied to monolayers of uninfected cells.
RT-PCR
IMR-90 cells were infected with VZV strain Ellen and total infected cell RNA was prepared at 48 hours post-infection using the RNAqueous-4PCR kit (Ambion, Inc., Austin, TX). RT-PCR was performed on RNA samples using the SuperScript One-Step RT-PCR kit with Platinum Taq (Invitrogen, Carlsbad, CA) according to the manufacturer's recommendations with the following primer pair: (ORF45 5′-CAAGTCTCGCCTGGAACAGT-3′, ORF42 5′-CAAGCTGTGACATCGCTATG-3′). The primer pairs were expected to yield a product of 386 nucleotides if ORF45 (Exon I) and ORF42 (Exon II) were contained within a spliced transcript. A synthetically fused ORF45/42 gene lacking an intron was used as a control template with the same primer set.
Cloning of Varicella-zoster virus strain Ellen open reading frames
Six of the seven VZV encapsidation ORFs (25, 26, 30, 34, 43, and 54) were amplified from VZV strain Ellen genomic DNA using gene specific oligonucleotide primers (IDT, Inc., Coralville, IA): ORF25: Forward 5′-CACCATGTACGAATCGGAAAATG-3′, Reverse 5′-AGCATCCTTCAATATTTCATG-3′; ORF26: Forward 5′-CACCATGGATCGGGTAGAATCAGA-3′, Reverse 5′-GACATACTTCGATAGGGTGTG-3′; ORF30: Forward 5′-CACCATGGAATTGGATATTAATCG-3′, Reverse 5′-TGAAAACGCCGGGTCCGTTG-3′; ORF34: Forward 5′-CACCATGACGGCGAGATATGGGTT-3′, Reverse 5′-CGGTGTGGAGGCAAACTGAGG-3′; ORF43: Forward 5′-CACCATGGAAGCCCATTTGGCAAAT-3′, Reverse 5′-TTTATGGGGGTTGGGAATAGAGAA-3′; ORF54: Forward 5′-CACCATGGCCGAAATAACGTCTC-3′, Reverse 5′-AGATCTTCGATCACGTCGC-3′. KOD HiFi DNA polymerase (TOYOBO/Novagen, EMD Biosciences Inc., Madison, WI) was used to generate blunt-end PCR products that were directionally cloned into pcDNA3.1D/V5-His-TOPO (Invitrogen). DNA sequencing (SeqWright, Houston, TX) confirmed that the V5 and His epitope tags were fused in frame after the last amino acid of each wild type, full length VZV ORF.
ORF 45/42 was cloned in several steps. Initially ORF45 (exon I) and ORF42 (exon II) were amplified from VZV strain Ellen genomic DNA using primers: ORF45: Forward 5′-CAAGAGATGAGGCGGATTCCGTGAATTG-3′; Reverse 5′-GCTGTAAGACCCGTCGGTTAACGAAAACGT; ORF42: Forward 5′-CGAGGTCAAGATGTTAACCTTCTGTTTGTGGA; Reverse 3′-AACCAGACTAGTTACATTTCACGCGTCTTG. Hpa I restriction enzyme sites (underlined) were incorporated into the ORF45 reverse and ORF42 forward primers by making silent mutations within the coding sequence. The ORF45 forward and ORF42 reverse primers were used to generate a single PCR product from a mixture of the individual ORF45 and ORF42 PCR products, which was subsequently cloned into pCR-Blunt-II-TOPO (Invitrogen). The resulting plasmid contained two Hpa I sites near the predicted splice junction of ORF45/42. Digestion with Hpa I removed unwanted sequences at the splice junction and two synthetic oligonucleotides (5′-AAAATCTTGACCTCGGATACCGTTTGTGTTGTGACTTGACGCGAACACCGCTGTGCTGTAAGACCCGTCGGTA-3′ and 5′-TACCGACGGGTCTTACAGCACAGCGGTGTTCGCGTCAAGTCACAACACAAACGGTATCCGAGGTCAAGATTTT-3′) were annealed, kinased and ligated into the Hpa I site. These sequences duplicated the splice junction predicted for wild type, full length ORF45/42. Full length, intron-less ORF45/42 was directionally cloned into pcDNA3.1D/V5-His-TOPO as described above using the primer pair: ORF45/42: Forward 5′-CACCATGTCATTGATAATGTTTGGTCG-3′; Reverse 5′-TTTAATAGGCATAAACACGGAATC-3′. DNA sequencing confirmed that the V5 epitope tag was fused in frame after the last amino acid of ORF42 and that no amino acid changes were introduced during regeneration of the predicted splice junction of ORF45/42.
Preparation of ORF30 and ORF45 specific antisera
Anti-ORF45/42 rabbit serum: Rabbits were immunized (New England Peptide, Inc., Gardner, MA) with a peptide corresponding to amino acids 19-33 (ERLKRRRDERFGTLE) from the N-terminus of the predicted amino acid sequence of ORF45. ELISA titers against the purified peptide were <50 and 912,900 for the pre-bleed and final bleed sera, respectively. Anti-ORF45/42 specific antibody was affinity purified via N-terminal peptide coupled column chromatography using the SulfoLink Kit (Pierce Biotechnology, Rockford, IL).
Anti-ORF30 guinea pig serum: Guinea pigs were immunized (Cocalico Biologicals, Inc., Reamstown, PA) with a recombinant GST fusion protein containing amino acids 307-577 of VZV ORF30.
Indirect immunofluorescence microscopy
Cells grown on sterile glass cover slips were either mock transfected or transfected with pcDNA3.1D/V5-ORF30, pcDNA3.1D/V5-ORF45/42, or pcDNA3.1D/V5-lacZ using Lipofectamine 2000 (Invitrogen). Forty-eight hours post-transfection, cells were fixed in a 50% methanol / 50% acetone solution, blocked in the presence of 3% bovine serum albumin (BSA), washed with phosphate buffered saline (PBS), and incubated with anti-V5 monoclonal antibody (Serotec, Inc., Raleigh, NC) for 1 hour. Subsequently, cover slips were washed with PBS containing 1% Triton X-100, incubated with fluorescein isothiocyanate-conjugated goat anti-mouse secondary antibody for 30 minutes, treated with 4′, 6-diamidino-2-phenylindole (DAPI), and mounted for examination by fluorescence microscopy.
Alternatively, glass cover slips were incubated for 24-48 hours with a mixture of uninfected and VZV strain Ellen infected cells. Cover slips prepared as described above were incubated with either an affinity purified anti-ORF45/42 rabbit serum, anti-ORF30 guinea pig serum, or a monoclonal antibody specific for the VZV major capsid protein (Virusys Corp., Sykesville, MD) followed by incubation with fluorescein isothiocyanate-conjugated goat anti-rabbit, anti-guinea pig, or anti-mouse secondary antibodies, respectively.
In vitro transcription/translation
In vitro transcription/translation of the pcDNA3.1D/V5-His-TOPO VZV ORF (Invitrogen) constructs was performed using a T7 coupled reticulocyte lysate system (Promega, Madison, WI). Approximately 2 µg of the plasmid was used for each 50 uL reaction as specified by the manufacturer. The in vitro products, containing the V5 epitope, were detected using an anti-V5 monoclonal antibody.
Immunoblotting
Proteins were detected by SDS-PAGE and immunoblot analysis. Transfected or in vitro translated products containing the V5 epitope were detected with an anti-V5 monoclonal antibody (1:5000) (Serotec, Inc.) followed by an anti-mouse HRP conjugated secondary antibody (1:3333). Infected cell proteins were detected using an anti-ORF30 guinea pig serum (1:1000) or a VZV anti-glycoprotein I (Chemicon, Temecula, CA) monoclonal antibody (1:5000) followed by anti-guinea pig (1:5000) or anti-mouse (1:3333) HRP conjugated secondary antibodies respectively. GST fusion proteins were detected with an anti-GST monoclonal antibody (1:5000) (EMD Biosciences Inc.) followed by an anti-mouse alkaline phosphatase conjugated secondary antibody (1:1000). Chemiluminescent detection was performed using the SuperSignal West Pico Chemiluminescent Substrate System (Pierce).
GST fusion protein purification
A DNA fragment containing the 3′ region of VZV ORF45/42 (bp1438-2241) was cloned into pGEX-3X (GE Healthcare, Piscataway, NJ). The resulting plasmid encoded a recombinant GST fusion protein, GST-ORF42-C269, containing the C-terminal 269 amino acids of ORF45/42, and represented 68% (269 out of 395 residues) of the full length ORF42-encoded exon II.
E. coli (JM109) transformed with pGEX-3X (GST) or pGEX-ORF42 (exon II) were inoculated into 200 ml of 2x-YT media containing 50 ug/mL ampicillin. Cultures were incubated at 30°C for eight hours and induced with 0.1 mM isopropyl-β-d-galactoside (IPTG) for 16 hours. Cells were harvested by centrifugation at 7,000 xg for 10 minutes, washed twice with PBS, re-suspended in lysis buffer (1.5% sarkosyl, 50 mg/mL lysozyme in PBS containing protease inhibitor cocktail) and sonicated briefly. Lysates were cleared by centrifugation at 10,000 xg for 5 minutes at 4°C. Supernatants were adjusted to 2% Triton-X 100, incubated at 4°C for one hour and purified on Sepharose 4B glutathione beads (GE Healthcare). Beads were washed five times for 10 minutes each with cold PBS and re-suspended in storage buffer (50 mM Hepes, pH 7.4, 150 mM NaCl, 5 mM dithiothreitol (DTT), 10% (v/v) glycerol) containing protease inhibitors. Proteins were fractionated by SDS-PAGE and analyzed for purity and integrity by Coomassie blue staining.
Additionally, a DNA fragment containing nucleotides 921-1731 of VZV ORF30 was cloned into pGEX-3X. The resulting plasmid expressed a GST fusion protein containing amino acids 307-577 of ORF30 which was purified from E. coli as described above and used as the antigen for the production of an ORF30 specific guinea pig antiserum.
GST-pull down assays
GST or GST-ORF42-C269 fusion protein immobilized on beads were removed from storage buffer and washed twice with binding (Proux-Gillardeaux et al., 2003) buffer (50 mM Hepes pH 7.4, 150 mM NaCl, 5 mM DTT, 10% v/v glycerol, 1% Triton X-100). Equal amounts of bead-immobilized GST or GST-ORF42 were re-suspended in 480 mL of binding buffer to which 20 uL of various V5-tagged in vitro transcribed / translated protein was added. After mixing at room temperature overnight, beads were washed five times for 10 min with binding buffer, solubilized in 2X sample buffer, fractionated on SDS-polyacrylamide gels and transferred to nitrocellulose membrane. Western blot analysis was performed using an anti-V5 monoclonal antibody to detect potential interaction of the in vitro synthesized gene products with the GST-ORF42-C269 fusion protein or the GST only control.
Subcellular fractionation
Mock or VZV strain Ellen infected MeWo cells were separated into their cytosolic and nuclear fractions using the Qproteome Cell Compartment kit (Qiagen Inc., Valencia, CA). The proteins within each fraction were precipitated with ice-cold acetone, dissolved in sample buffer, and resolved by SDS-PAGE. Gels were either stained with Coomassie blue or used in immunoblot analysis.
Results
Cloning of the VZV strain Ellen open reading frames encoding the putative DNA cleavage and packaging gene homologs
Based on sequence homology, seven VZV genes, ORFs 25, 26, 30, 34, 43, 45/42, and 54, were identified as homologs of the HSV-1 DNA encapsidation genes. The entire coding region for six of the seven ORFs was amplified from VZV strain Ellen genomic DNA. ORFs 45 and 42 were the most likely candidates to encode exon I and exon II, respectively, of a spliced gene product conserved in both HSV-1 (UL15) and HCMV (UL89). ORFs 45 and 42 were amplified separately and fused to yield a continuous open reading frame lacking the putative intron sequences derived from ORFs 43 and 44.
Characteristics for all seven of the VZV Ellen encapsidation gene homologs are presented in Table I. The predicted amino acid sequences of the Ellen ORFs were compared to those of nineteen other previously published VZV isolates. Not unexpectedly, few amino acid substitutions were observed. There were no differences in ORF25 and ORF30 for any of the isolates. The small number of differences observed for ORF34, ORF54 and ORF45/42 were all conservative in nature. Only three non-conservative substitutions were observed: 853 Arg>Cys of ORF26 in a set of three related isolates (32p72, 33p22, 32p5), 177 Arg>Cys of ORF43 for the MSP isolate, and 466 Pro>Leu of ORF43 for all nineteen isolates. The biological significance of these changes, if any, is not known. Overall, a high degree of conservation existed at both the nucleotide and amino acid levels for the 20 isolates that were compared.
Table 1.
Sequence analysis of the VZV strain Ellen DNA encapsidation genes, ORFs 25, 26, 30, 34, 43, 45/42, and 54. ORF 45/42 is expressed in VZV-infected cells as a spliced gene product derived from two exons (ORF42 and ORF45) (see Fig. 1). The column labeled “amino acid change” represents the amino acid differences observed for one or more of the nineteen published VZV isolates compared to Ellen. The predicted molecular mass of each protein product with and without the V5 epitope tag is indicated. The V5 tag is estimated to add 5.1 kDa to each wild type protein.
| ORF | GenBank
Accession |
bp | aa | kDa | (+ V5) | Isolates with Amino Acid Differences
Compared to VZV Strain Ellena |
Amino Acid
Change |
|---|---|---|---|---|---|---|---|
| 25 | DQ145725 | 468 | 156 | 17.5 | 22.6 | None | *** |
| 26 | DQ077821 | 1755 | 585 | 65.7 | 70.8 | 32p5, 32p22, 32p72 | 285 R>C |
| 30 | DQ075209 | 2310 | 770 | 87.0 | 92.1 | None | *** |
| 34 | DQ077822 | 1737 | 579 | 65.2 | 70.3 | 36, 49, BC | 155 T>A |
| 43 | DQ531049 | 2028 | 676 | 73.9 | 79.0 | 8, 36, 49, 32p5, 32p22, 32p72, BC, Dumas, Kel, MSP, | 126 I>V |
| Oka Parent, Oka Vaccine, SD, VarilRix, VariVax | |||||||
| MSP | 177 R>C | ||||||
| 8 | 222 I>T | ||||||
| 36, 49 | 323 A>G | ||||||
| All | 466 P>L | ||||||
| 45/42 | DQ910905 | 2241 | 747 | 82.8 | 87.9 | All | 247 M>T |
| 54 | DQ910906 | 2307 | 769 | 86.8 | 91.2 | VarilRix, VariVax, Oka Parental, Oka Vaccine | 128 E>D |
| HJO | 711 V>I |
Isolates : 03-500, 8, 11, 22, 36, 49, 32p5, 32p22, 32p72, BC, Dumas, HJO, Kel, MSP, Oka Parent, Oka Vaccine, SD, VarilRix, VariVax
RT-PCR analysis of total RNA isolated from VZV-infected cells reveals a spliced ORF45/42 gene product
The UL15 gene of herpes simplex viruses consists of two coding exons separated by an intron of 3587 bp (Baines and Roizman, 1992; Dolan et al., 1991). The intron contains two genes, UL16 and UL17, transcribed in the opposite orientation of UL15. Based on sequence homology, ORF45 and ORF42 would be the most likely candidates to encode an equivalent spliced gene product in VZV-infected cells (Fig. 1A) (Davison and Scott, 1986). RT-PCR analysis was performed on RNA isolated from VZV strain Ellen infected IMR-90 cells (Fig. 1A). Primers specific for the 3′ end of exon I (ORF45) and the 5′ end of exon II (ORF42) resulted in the synthesis of a 386 bp product that was cloned, sequenced and confirmed to be the spliced product of the two predicted exons. An identical fragment was observed when using the same two primers with a synthetically spliced ORF45/42 template. No similar products were observed in RNA prepared from uninfected cells. Sequencing of the 386 bp product confirmed the splice junction and that exon I (ORF45) and exon II (ORF42) encoded 1056 and 1185 nucleotides respectively. The results suggested that ORF45/42 is expressed as a spliced product consistent with the previously published results for the UL15 gene.
Fig. 1.
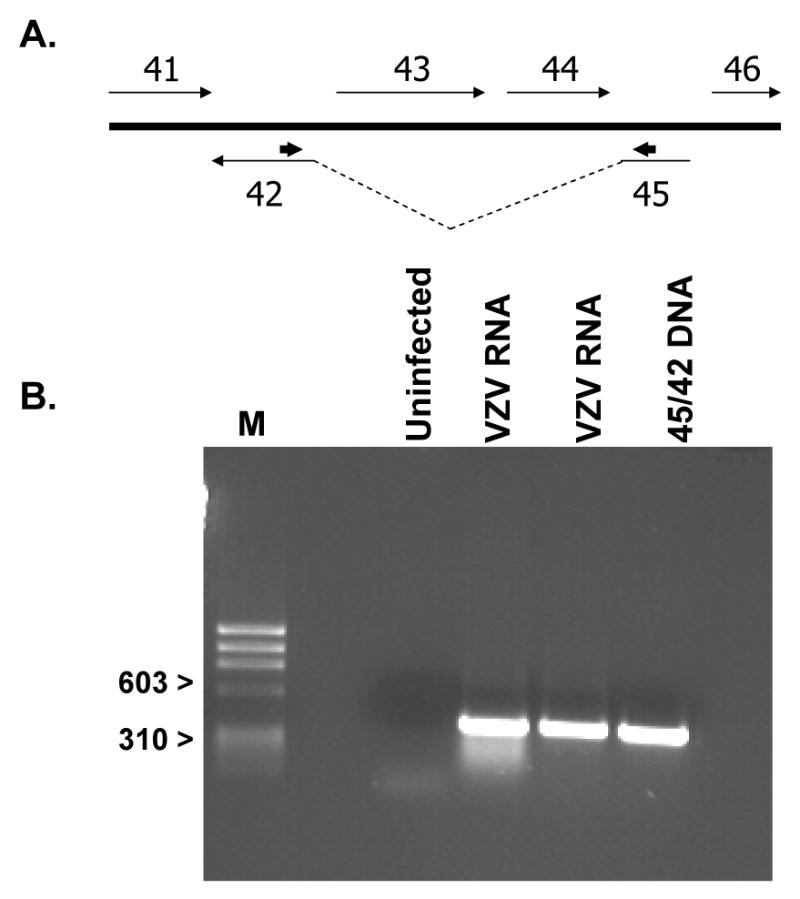
Identification of the spliced ORF45/42 RNA transcript. (A) Arrangement of the ORFs in VZV showing the location of ORFs 45 and 42 and the intervening sequences encoded by ORFs 43 and 44. The direction of transcription is indicated by the thin arrows. The approximate location of the forward and reverse primers used for RT-PCR is shown by the short arrows (->) above the 3′-ORF45 and 5′-ORF42 sequences. (B) RT-PCR analysis of RNA samples prepared from VZV strain Ellen infected IMR-90 cells. The primer pair was expected to yield a fragment of 386-bp if a spliced ORF45/42 product was present. RNA was isolated from uninfected cells (Neg) and two different VZV-infected cell samples. Additionally, a DNA fragment representing the expected product was generated via PCR using a synthetically constructed ORF45/42 plasmid (45/42 DNA) lacking the intervening sequences. Markers of 310 and 603 bp are shown on the left (M).
In vitro expression of epitope tagged VZV encapsidation gene products
Each of the open reading frames was cloned into pcDNA3.1D/V5-His-TOPO and protein products were expressed via coupled in vitro transcription and translation. The resulting products were analyzed by SDS-PAGE followed by immunoblotting with an antibody that recognized a C-terminal V5 epitope fused in frame with the VZV ORF (Fig. 2). The molecular mass of the major polypeptide observed in each sample was consistent with that predicted for each respective ORF (Table 1). (The V5 epitope tag added approximately 5.1 kDa to each full length VZV protein.)
Fig. 2.
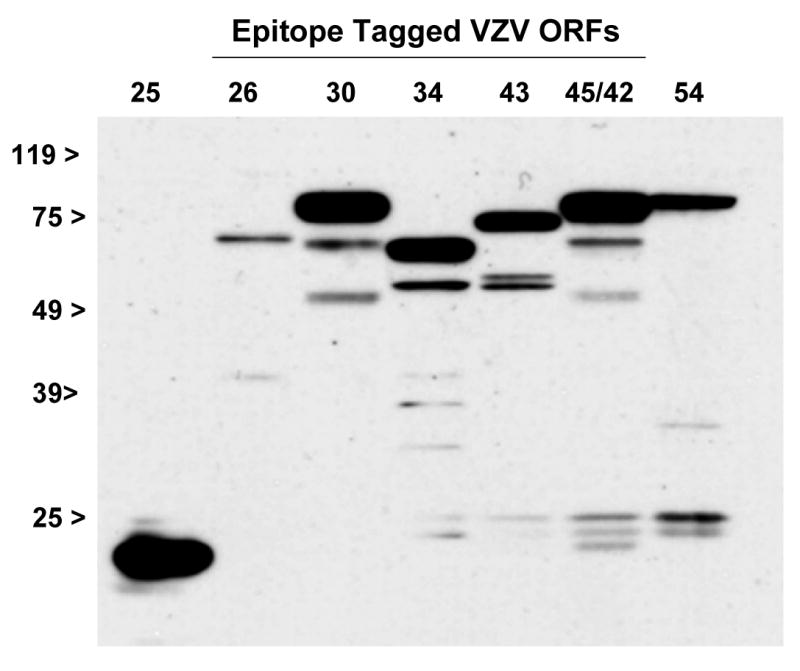
The DNA encapsidation genes, ORFs 25, 26, 30, 34, 43, 45/42, and 54, were cloned into pcDNA3.1D/V5-His-TOPO. In vitro transcription and translation was used to synthesize C-terminal V5 epitope tagged polypeptides. Proteins were detected using an anti-V5 monoclonal antibody. The predicted molecular mass for each protein product with and without the V5 epitope is provided in Table I.
Expression of the putative ORF45/42 terminase subunit in transfected and VZV-infected cells
The sub-cellular localization of ORF45/42-V5 was examined via indirect immunofluorescence microscopy. Using an anti-V5 antibody, ORF45/42-V5 was shown to localize primarily to the cytoplasm of transfected MeWo cells (Fig. 3A). Weak nuclear staining was observed in some transfected cells but none showed strong or exclusive nuclear staining. In comparison, the HSV-1 homolog of ORF45/42, UL15, was reported to localize to the nucleus of transfected cells (Koslowski et al., 1999; Yu and Weller, 1998a). Differences in localization might be explained by the presence of a strong nuclear localization signal in the UL15 polypeptide (PKKRAKV at amino acids 183-189) but only a partially homologous sequence predicted for ORF45/42 (KRAKV at amino acids 194-198).
Fig. 3.
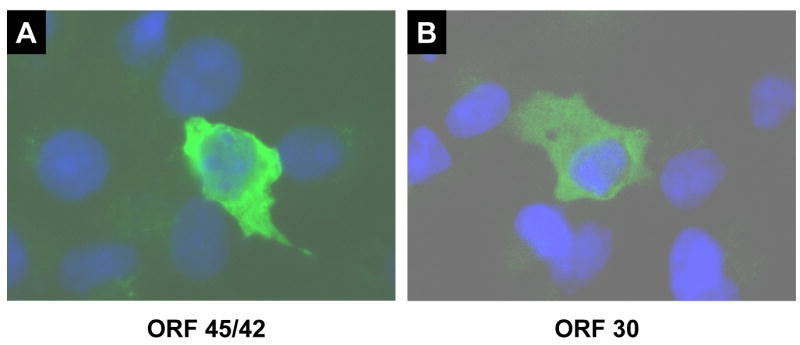
Indirect immunofluorescence analysis of ORF45/42 and ORF30 expressed in transiently transfected cells. Cells were mock transfected or transfected with pcDNA3.1D/V5-ORF45/42 or pcDNA3.1D/V5-ORF30. Twenty-four hours post-transfection cells were fixed, stained with DAPI, and incubated with an anti-V5 primary antibody followed by a FITC conjugated secondary antibody.
The cellular localization of the ORF45/42 gene product in the context of VZV infection was examined. Coverslips containing uninfected cells (Fig. 4A) or a mixture of uninfected and VZV-infected MRC5 (Fig. 4B), HFF (Fig. 4C and D), or MeWo (Fig. 4E-H) cells were fixed with ice-cold acetone at 48 hours post-infection and incubated with an affinity purified anti-ORF45/42 peptide rabbit serum (Fig. 4A-F) or a monoclonal antibody to the VZV major capsid protein (Fig. 4G and H). ORF45/42 showed distinct nuclear staining in all three cell lines (Fig. 4B, D, and F) similar to that observed for the major capsid protein (Fig. 4H). Uninfected cells (Fig. 4A) and cells incubated with pre-immune serum (data not shown) were negative for fluorescence. In contrast to the primarily cytoplasmic localization of ORF45/42-V5 in transfected cells, the ORF45/42 gene product was expressed almost exclusively in the nucleus of VZV-infected cells. The results suggest that one or more viral gene products might be involved in the translocation of ORF45/42 into the nucleus of infected cells.
Fig. 4.
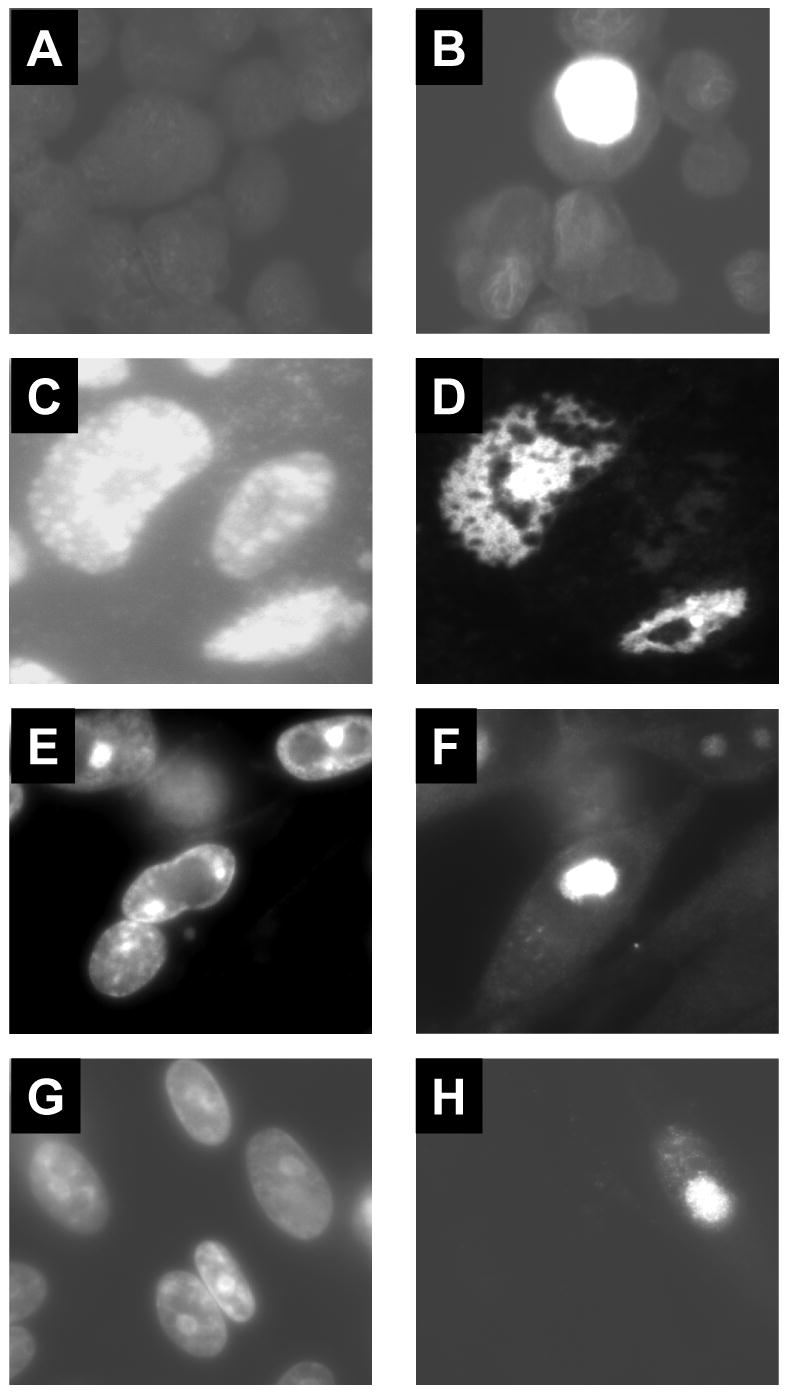
Indirect immunofluorescence analysis of VZV-infected cells. Cover slips were prepared with uninfected MRC5 (A) or a mixture of uninfected and VZV-infected MRC5 (B), HFF (C and D) or MeWo (E-H) cells. Cells were incubated with an affinity purified anti-ORF45/42 peptide serum (A-F) or a monoclonal antibody specific for the VZV major capsid protein (G and H)). Panels C, E, and G show the adjacent field of cells stained with DAPI.
The peptide antiserum reacted only weakly in immunoblot analysis of in vitro synthesized ORF45/42-V5 (data not shown). Probing of VZV-infected cell extracts failed to consistently detect polypeptides that could be assigned as ORF45/42 specific.
Expression of the putative ORF30 terminase subunit in transfected or VZV-infected cells
The sub-cellular localization of ORF30-V5 was examined via indirect immunofluorescence. Using an anti-V5 antibody, ORF30-V5 was shown to localize primarily to the cytoplasm of transfected MeWo cells (Fig. 3B). Little to no nuclear staining was observed in transfected cells. The results were consistent with the previously reported observation that the HSV-1 homolog of ORF30, UL28, localized to the cytoplasm of transfected cells (Koslowski et al., 1999; Koslowski et al., 1997).
The cellular localization of the ORF30 gene product in the context of VZV infection was examined. In contrast to the ORF45/42 gene product, an anti-ORF30 guinea pig serum showed varied localization of ORF30 including cytoplasmic and perinuclear staining (data not shown). Immunoblot analysis using the anti-ORF30 serum readily detected an 87 kDa polypeptide in VZV-infected cells but not in uninfected cell extracts (Fig. 5). This size was consistent with that predicted for the full length ORF30 protein (Table 1). Based on sequence homology to other herpesviral terminase subunits (Visalli and van Zeijl, 2003), ORF30 likely encodes one of the two putative VZV terminase subunits. Since DNA encapsidation occurs in the nucleus of herpsevirus infected cells, it was hypothesized that some ORF30 would be observed in the nucleus of VZV infected cells. Therefore, mock or VZV strain Ellen infected MeWo cells were separated into their cytosolic and nuclear fractions. Proteins in each fraction were analyzed by SDS-PAGE and Coomassie blue staining. The presence of the 155 kDa VZV major capsid protein was used to monitor the fractionation procedure since the major capsid protein is a primary component of virions and known to localize to the nucleus of infected cells (Kinchington et al., 1992; also see Fig. 4H this study). The 155 kDa protein was readily visible in both the nuclear fraction of infected cells and total infected cell extracts but not in the cytoplasmic fraction or mock infected cells extracts (Fig. 6A). When the same samples were used in immunoblot analysis with anti-ORF30 serum, the 87 kDa ORF30 gene product co-fractionated with the major capsid protein (Fig. 6B) found in the nuclear fraction of cell extracts. Some ORF30 was also detected in the cytosolic fraction and this is consistent with reports that encapsidation proteins, including the terminase subunits of HSV-1, can be detected in the cytoplasm of infected cells (Yang et al., 2007). Further studies will be necessary to define the role of ORF30 in VZV encapsdiation, however, the presence of ORF30 in the nucleus of infected cells is consistent with that of a terminase subunit.
Fig. 5.
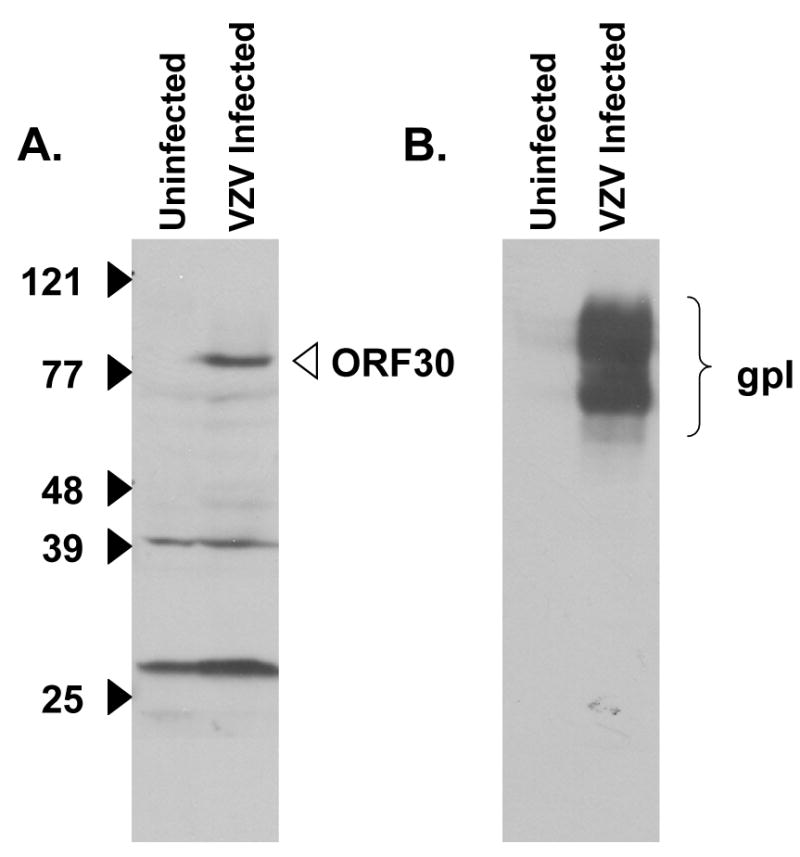
Detection of the VZV ORF30 protein in VZV-infected MeWo cells. Immunoblot analysis of uninfected or VZV Ellen infected cell extracts was performed with (A) an anti-ORF30 guinea pig serum or (B) a monoclonal antibody specific for VZV glycoprotein I (a positive control for infected cells proteins).
Fig. 6.
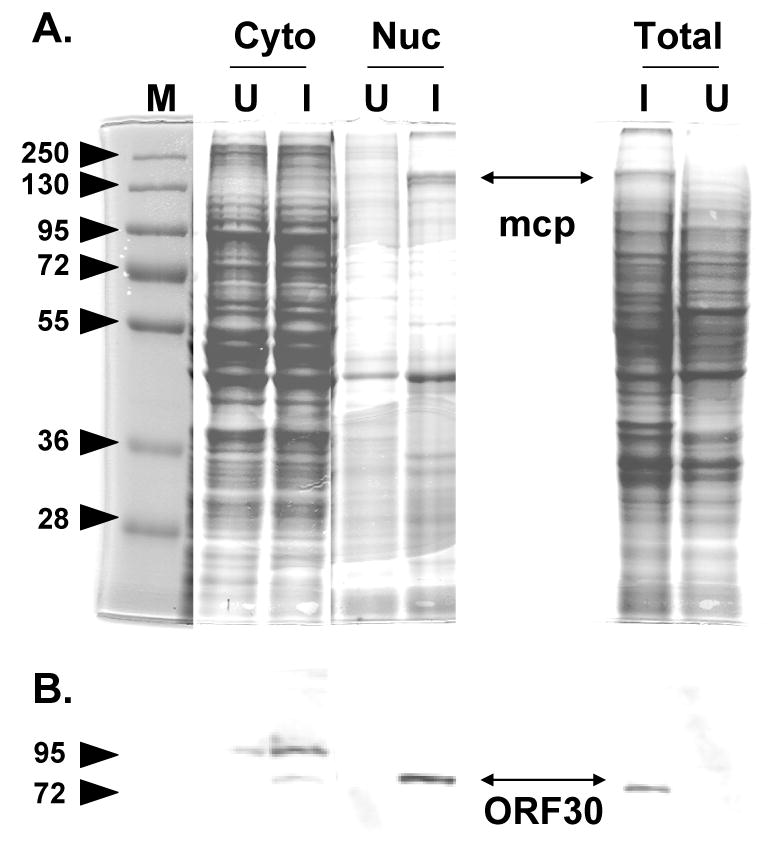
Subcellular fractionation of mock infected and VZV infected cells. (A) Coomassie stained gel of the cytoplasmic (cyto) or nuclear (nuc) fractions of uninfected (U) or infected (I) cell extracts. The location of the 155 kDa VZV major capsid protein is shown in the infected cell nuclear fraction and total infected cell extract. (B) Immublot analysis using anti-ORF30 serum with a duplicate set of samples observed in panel A. The 87 kDa ORF30 protein is indicated. mcp = major capsid protein.
Interaction of the VZV terminase subunits
Studies with HSV-1 and HCMV defined several interactions between the terminase subunits of the respective viruses. The human cytomegalovirus subunits, pUL56 and pUL89, were shown to interact by both co-immunoprecipitation (Hwang and Bogner, 2002) from infected cell extracts and in vitro GST pull down assays. Results from pull down assays using GST-UL89 fusion proteins and in vitro synthesized pUL56 suggested that C-terminal residues of pUL89 were necessary for the interaction of the terminase subunits (Thoma et al., 2006). A fusion protein consisting of the carboxyl-terminal portion of pUL56 was shown to interact with full length in vitro synthesized pUL89 (Hwang and Bogner, 2002). The HSV-1 subunits, UL28 and UL15, were shown to interact via co-immunoprecipitation of baculovirus expressed UL28 and UL15 polypeptides (Abbotts et al., 2000). The results suggested that several regions of UL28 interacted with UL15 and also that the C-terminus of UL15 (UL15-exon II) could mediate interaction with the full length UL15.
Pull downs assay were performed to determine if similar interactions could be observed for the VZV ORF45/42 and ORF30 gene products. V5 epitope tagged ORF30, ORF45/42, or β-gal (β-galactosidase) polypeptides were incubated with purified GST or GST-ORF42-C269 immobilized on glutathione beads. Western blot analysis was performed on each pull down assay to detect potential interactions between the in vitro synthesized products and GST-ORF42-C269 (Fig. 7C). The 45/42-V5 and 30-V5 polypeptides were observed to interact with GST-ORF42-C269 (Fig. 7C, lanes 1 and 5) but not with GST alone (Fig. 7C, lanes 2 and 6). β-gal-V5 did not interact with GST or GST-ORF42-C269 (Fig. 7C, lanes 4 and 3). The results showed that the C-terminal 269 amino acids of exon II (ORF42) were sufficient to mediate interaction with the full length ORF45/42 or ORF30 gene products. Furthermore, the results suggested that the ORF45/42 gene product has the potential to form complexes with itself via interaction through exon II. These results are consistent with those observed for the HSV-1 terminase subunits.
Fig. 7.
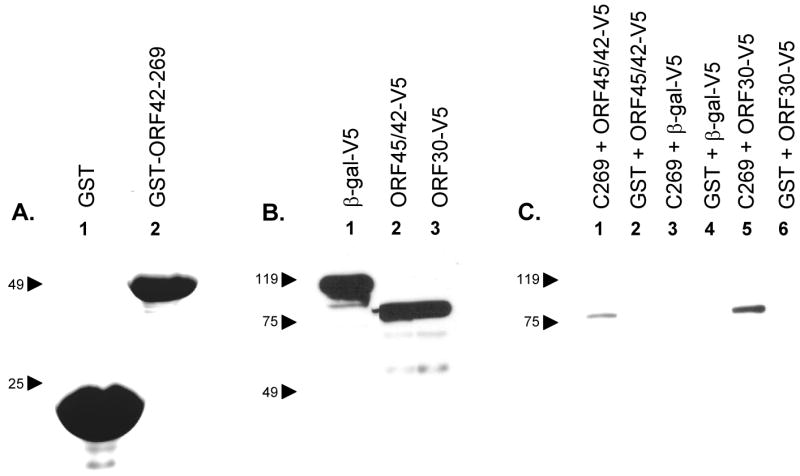
GST pull down assays were performed to detect potential protein-protein interactions. (A) GST (lane 1) or GST-ORF42-C269 (lane 2) were purified on glutathione beads. Proteins were analyzed on Coomassie blue stained SDS-polyacrylamide gels. (B) pcDNA3.1D/V5 - lacZ (β-gal; lane 1), ORF45/42 (lane 2), or ORF30 (lane 3) were used in in vitro transcription/translation reactions to synthesize the respective protein products containing a V5 epitope. (C) GST (lanes 2, 4, and 6) or GST-ORF42-C269 (lanes 1, 3, and 5) were immobilized on beads and re-suspended in binding buffer containing ORF45/42-V5 (lanes 1 and 2), β-gal-V5 (lanes 3 and 4) or ORF30-V5 (lanes 5 and 6). Beads were washed, fractionated by SDS-PAGE and subjected to immunoblot analysis using an anti-V5 monoclonal antibody.
Affinity of the ORF30 and ORF45/42 interactions with ORF42-C269
The original pull down assay that showed an interaction between the C-terminus of ORF45/42 and ORF30 was performed under low ionic conditions (150 mM NaCl) in the presence of 1% Triton X-100 (a non-ionic detergent). To show that this interaction could be maintained under more stringent conditions, pull down assays were performed in the presence of increasing concentrations of (i) anionic detergents (Fig. 8) or (ii) NaCl (data not shown).
Fig. 8.
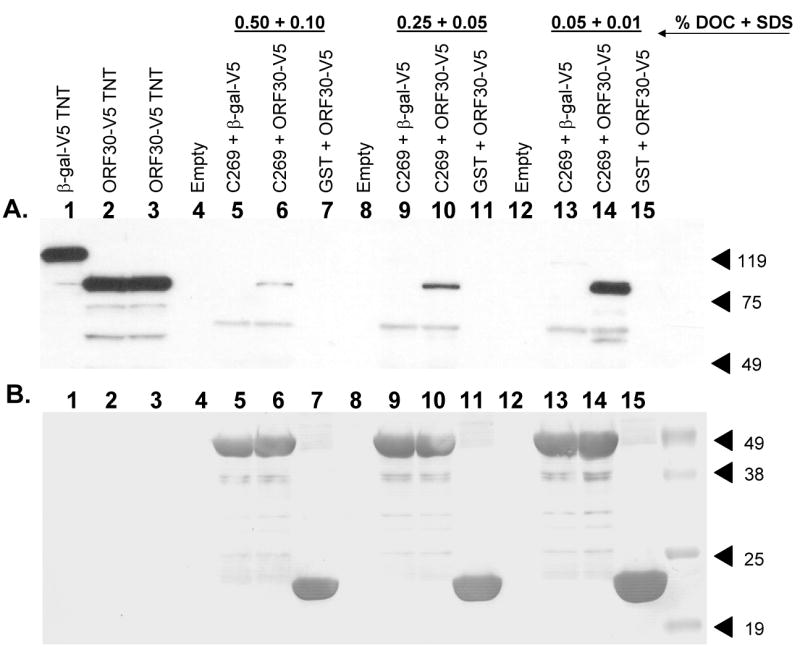
Effect of varying anionic detergent concentrations on the binding of ORF30 to ORF42-C269. β-gal-V5 (lane 1) or ORF30-V5 (lanes 2 and 3) were incubated with purified GST or GST-ORF42-C269 in the presence of 0.05% DOC + 0.01% SDS (lanes 13-15), 0.25% DOC + 0.05% SDS (lanes 9-11) or 0.50% DOC + 0.10% SDS (lanes 5-7). Samples were subjected to immunoblotting first with an anti-V5 monoclonal antibody (A) and after stripping the blot, with an anti-GST monoclonal antibody (B). Lanes: (1) β-gal-V5; (2-3) ORF30-V5; (4, 8 and 12) empty; (5, 9 and 13) GST-ORF42-C269 + β-gal-V5; (6, 10 and 14) GST-ORF42-C269 + ORF30-V5; (7, 11 and 15) GST + ORF30-V5. (Lanes 4, 8, and 12 were empty.)
Pull down assays were performed as described previously except that the binding and wash buffers contained increasing concentrations of the anionic detergents, SDS and DOC [0.05% DOC + 0.01% SDS (lanes 13-15), 0.25% DOC + 0.05% SDS (lanes 9-11) or 0.50% DOC + 0.10% SDS (lanes 5-7)]. Interactions were maintained at the highest concentrations tested (Fig. 8A, lane 6). A decrease in relative binding was observed as the detergent concentrations increased (Fig. 8A, compare lanes 14, 10, and 6). β-gal-V5 did not bind GST-ORF42-C269 (Fig. 8a, lanes 5, 9, and 13), nor did ORF30 bind GST (Fig 8A., lanes 7, 11, and 15), for any of the conditions tested. Stripping and reprobing the blot with an anti-GST monoclonal antibody (Fig. 8B) showed that all lanes contained approximately equal amounts of GST (Fig. 8B, lanes 7, 11, and 15) or GST-ORF42-C269 fusion protein (Fig. 8B, lanes 5-6, 9-10, and 13-14) in the binding assays.
Pull down assays were performed as described previously except that the binding and wash buffers contained increasing concentrations of NaCl (50 mM NaCl, 150 mM NaCl, 250 mM NaCl, or 500 mM NaCl). Interactions were maintained under conditions of increasing ionic strength, however, a decrease in relative binding was observed as the ionic strength increased (data not shown). The β-gal protein did not bind GST-ORF42-C269 nor did ORF30-V5 bind GST for any of the conditions tested.
Interaction between ORF30 and ORF42-C269 was observed under increasingly stringent conditions that were maintained during both the binding and washing steps of the pull down assays. The results are consistent with previous studies demonstrating interaction between the terminase subunits of other herpesviruses.
Discussion
Previous studies performed on the DNA encapsidation proteins of HSV-1, and to a lesser extent on HCMV, suggest that certain interactions are conserved between homologous encapsidation proteins of the family Herpesviridae. Studies designed to elucidate interactions amongst the VZV DNA encapsidation proteins will contribute to a more thorough understanding of the protein-protein complexes that take part, and are likely conserved, in the process of herpesviral DNA cleavage and packaging. The results presented in this paper are the first that pertain to any of the putative VZV encapsidation protein homologs. More specifically, in vitro pull down assays revealed that the putative VZV terminase subunits, encoded by ORF30 and ORF45/42, interacted with each other via the second exon of ORF45/42. Additionally, the C-terminal 269 amino acids of ORF45/42-exon II were shown to be sufficient to mediate self-interaction with the full length ORF45/42 gene product.
These results are consistent with those previously described for the HSV-1 and HCMV terminase subunit homologs. The C-terminal portion of the HSV-1 ORF45/42 homolog, UL15, was sufficient to mediate interaction with the full length UL15 gene product. The terminase subunits of HSV-1 or HCMV were shown to interact via GST pull down assays, co-immunoprecipitation of baculovirus expressed products, and/or from virus infected cell extracts.
We showed the interaction of ORF30 or ORF45/42 with ORF42-C269 was maintained in increasing detergent concentrations and in the presence of increasing ionic strength. The results suggest that a relatively stable association occurs between the VZV terminase subunits. Confirmation of ORF30 and ORF45/42 interactions in VZV-infected cells has proven challenging. This might be attributed to the inherent difficulty of infecting cells with VZV at a high multiplicity and/or the possibility that terminase subunit complexes are present in low amounts in infected cells.
Prior to the studies reported here, none of the VZV encapsidation proteins had been identified in infected cells. The identification of a spliced mRNA consisting of two exons derived from ORFs 45 and 42 is analogous to that previously reported for the UL15 and UL89 genes of HSV-1 and HCMV respectively. An ORF45/42 specific peptide antiserum showed that the ORF45/42 gene product localized to the nucleus of VZV-infected cells. This result is consistent with ORF45/42 encoding a terminase involved in the DNA encapsidation process. Interestingly, cells transiently transfected with ORF45/42 showed only weak nuclear staining. This is in contrast to the HSV-1 homolog, UL15, which was shown to localize efficiently to the nucleus in UL15 transfected cells. This is not entirely unexpected as UL15 contains a readily identifiable nuclear localization signal whereas ORF45/42 does not. Further studies will be necessary to determine if another VZV protein, cell protein, and/or modification of ORF45/42 is required for the translocation of the ORF45/42 gene product to the nucleus in infected cells.
In contrast to the striking nuclear localization observed for the ORF45/42 gene product, localization of the ORF30 gene product by indirect immunofluorescence was less clear. Immunoblot analysis of VZV-infected cell extracts using an ORF30 specific antiserum readily detected a polypeptide of 87 kDa. Analysis of the nuclear fraction isolated from VZV infected cell extracts revealed co-fraction of ORF30 with the VZV major capsid protein. Sequence homology with other herpesvirus terminase subunit genes strongly suggested that the ORF30 gene product was one of the two putative VZV terminase subunits. Therefore, some ORF30 gene product would be expected to localize to the nucleus in infected cells.
Our results provide corroborating evidence that the DNA encapsidation proteins of the Herpesvirdae form a heterodimeric terminase. Several classes of herpesviral inhibitors have been described that target the viral terminase complex (Buerger et al., 2001; Krosky et al., 1998; Reefschlaeger et al., 2001). Interestingly, there appear to be separate classes of inhibitors that act upon different viral cleavage and packaging proteins associated with the terminase complex.
The benzimidazole compounds BDCRB and TCRB were shown to be potent and highly specific inhibitors for the terminase complex of HCMV (Townsend et al., 1995). Treatment of HCMV infected cells with either resulted in the accumulation of immature capsids and uncleaved DNA (Krosky et al., 1998). Maximal resistance to these compounds was mapped to two different encapsidation genes, UL89 and UL56 (Krosky et al., 1998; Underwood et al., 1998). The UL89/56 gene products are thought to make up the heterodimeric HCMV terminase complex.
Buerger et al. (2001) described a series of non-nucleoside inhibitors (BAY 38-4766 and associated analogs) that interfere with HCMV viral DNA cleavage and packaging via the UL89 and UL56 terminase subunits but whose molecular mode of interaction is in fact separable from the benzimidazole ribonucleosides. This BAY series of compounds differs from the previously reported HCMV terminase inhibitors in i) the apparent lack of demonstrable viral cross-resistance to the benzimidazole ribonucleoside series (Evers et al., 2002) and ii) their broadened range of specificity (i.e. they also inhibit monkey and rodent cytomegaloviruses) (Buerger et al., 2001; Reefschlaeger et al., 2001; Weber et al., 2001).
Protein-protein interactions found to occur between the encapsidation proteins of one member of the Herpesviridae may be conserved in one or more of the other family members. The understanding of such interactions could be utilized to develop assays for the identification of new small molecular inhibitors of DNA encapsidation. The discovery of small molecules that inhibit DNA cleavage or packaging for HSV-1, HCMV, and VZV validate encapsidation as an antiviral target (Biron et al., 2002; van Zeijl et al., 2000; Visalli et al., 2003; Visalli and van Zeijl, 2003). The reagents described in this report, combined with a series of VZV specific small molecule encapsidation inhibitors that target the putative ORF54 portal protein (Visalli et al., 2003; Di Grandi et al., 2004), should be valuable tools to study the multi-protein complexes involved in DNA cleavage and packaging. Further dissection of the interactions between the ORF30 and ORF45/42 gene products and investigation of potential interactions between the other VZV encapsidation proteins merits further study.
Acknowledgments
We would like to thank Dr. Charles Grose (University of Iowa) for providing MeWo cell stocks. These studies were supported by an American Society for Microbiology Undergraduate Summer Research Grant (to D.M.N.) and National Institutes of Health grant 1 R15 AI062713-01.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Abbotts AP, Preston VG, Hughes M, Patel AH, Stow ND. Interaction of the herpes simplex virus type 1 packaging protein UL15 with full-length and deleted forms of the UL28 protein. J Gen Virol. 2000;81(Pt 12):2999–3009. doi: 10.1099/0022-1317-81-12-2999. [DOI] [PubMed] [Google Scholar]
- Alba MM, Das R, Orengo CA, Kellam P. Genomewide function conservation and phylogeny in the Herpesviridae. Genome Res. 2001;11(1):43–54. doi: 10.1101/gr.149801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baines JD, Roizman B. The cDNA of UL15, a highly conserved herpes simplex virus 1 gene, effectively replaces the two exons of the wild-type virus. J Virol. 1992;66(9):5621–6. doi: 10.1128/jvi.66.9.5621-5626.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beard PM, Taus NS, Baines JD. DNA cleavage and packaging proteins encoded by genes U(L)28, U(L)15, and U(L)33 of herpes simplex virus type 1 form a complex in infected cells. J Virol. 2002;76(10):4785–91. doi: 10.1128/JVI.76.10.4785-4791.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biron KK. Antiviral drugs for cytomegalovirus diseases. Antiviral Res. 2006;71(23):154–63. doi: 10.1016/j.antiviral.2006.05.002. [DOI] [PubMed] [Google Scholar]
- Biron KK, Harvey RJ, Chamberlain SC, Good SS, Smith IA, Davis MG, Talarico CL, Miller WH, Ferris R, Dornsife RE, Stanat SC, Drach JC, Townsend LB, Koszalka GW. Potent and Selective Inhibition of Human Cytomegalovirus Replication by 1263W94, a Benzimidazole L-Riboside with a Unique Mode of Action. Antimicrob Agents Chemother. 2002;46(8):2365–72. doi: 10.1128/AAC.46.8.2365-2372.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bogner E. Human cytomegalovirus terminase as a target for antiviral chemotherapy. Rev Med Virol. 2002;12(2):115–27. doi: 10.1002/rmv.344. [DOI] [PubMed] [Google Scholar]
- Bogner E, Radsak K, Stinski MF. The gene product of human cytomegalovirus open reading frame UL56 binds the pac motif and has specific nuclease activity. J Virol. 1998;72(3):2259–64. doi: 10.1128/jvi.72.3.2259-2264.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown J, M MA, Homa FL. Packaging DNA into herpesvirus capsids. In: Holzenburg B, editor. Structure-function relationships of human pathogenic viruses. Kluwer academic/Plenum; New York: 2002. pp. 111–153. [Google Scholar]
- Buerger I, Reefschlaeger J, Bender W, Eckenberg P, Popp A, Weber O, Graeper S, Klenk HD, Ruebsamen-Waigmann H, Hallenberger S. A novel nonnucleoside inhibitor specifically targets cytomegalovirus DNA maturation via the UL89 and UL56 gene products. J Virol. 2001;75(19):9077–86. doi: 10.1128/JVI.75.19.9077-9086.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casper C. Defining a role for antiviral drugs in the treatment of persons with HHV-8 infection. Herpes. 2006;13(2):42–7. [PubMed] [Google Scholar]
- Casper C, Wald A. The use of antiviral drugs in the prevention and treatment of Kaposi sarcoma, multicentric Castleman disease and primary effusion lymphoma. Curr Top Microbiol Immunol. 2007;312:289–307. doi: 10.1007/978-3-540-34344-8_11. [DOI] [PubMed] [Google Scholar]
- Chang YE, Poon AP, Roizman B. Properties of the protein encoded by the UL32 open reading frame of herpes simplex virus 1. J Virol. 1996;70(6):3938–46. doi: 10.1128/jvi.70.6.3938-3946.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davison AJ, Scott JE. The complete DNA sequence of varicella-zoster virus. J Gen Virol. 1986;67(Pt 9):1759–816. doi: 10.1099/0022-1317-67-9-1759. [DOI] [PubMed] [Google Scholar]
- De Clercq E. Antiviral drugs: current state of the art. J Clin Virol. 2001;22(1):73–89. doi: 10.1016/s1386-6532(01)00167-6. [DOI] [PubMed] [Google Scholar]
- Di Grandi MJ, Bloom JD, Curran KJ, Feigelson G, Prashad A, Ross AA, Visalli RJ, Feld B. Thiourea inhibitors of Herpesviruses Part 3: Inhibitors of VZV. Bioorg Med Chem Lett. 2004;16(14):4157–4160. doi: 10.1016/j.bmcl.2004.06.025. [DOI] [PubMed] [Google Scholar]
- Dittmer A, Drach JC, Townsend LB, Fischer A, Bogner E. Interaction of the putative human cytomegalovirus portal protein pUL104 with the large terminase subunit pUL56 and its inhibition by benzimidazole-D-ribonucleosides. J Virol. 2005;79(23):14660–7. doi: 10.1128/JVI.79.23.14660-14667.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dolan A, Arbuckle M, McGeoch DJ. Sequence analysis of the splice junction in the transcript of herpes simplex virus type 1 gene UL15. Virus Res. 1991;20(1):97–104. doi: 10.1016/0168-1702(91)90064-3. [DOI] [PubMed] [Google Scholar]
- Evers DL, Komazin G, Shin D, Hwang DD, Townsend LB, Drach JC. Interactions among antiviral drugs acting late in the replication cycle of human cytomegalovirus. Antiviral Res. 2002;56(1):61–72. doi: 10.1016/s0166-3542(02)00094-3. [DOI] [PubMed] [Google Scholar]
- Goshima F, Watanabe D, Takakuwa H, Wada K, Daikoku T, Yamada M, Nishiyama Y. Herpes simplex virus UL17 protein is associated with B capsids and colocalizes with ICP35 and VP5 in infected cells. Arch Virol. 2000;145(2):417–26. doi: 10.1007/s007050050033. [DOI] [PubMed] [Google Scholar]
- Griffiths PD. Antivirals in the transplant setting. Antiviral Res. 2006;71(23):192–200. doi: 10.1016/j.antiviral.2006.04.016. [DOI] [PubMed] [Google Scholar]
- Hwang JS, Bogner E. ATPase activity of the terminase subunit pUL56 of human cytomegalovirus. J Biol Chem. 2002;277(9):6943–8. doi: 10.1074/jbc.M108984200. [DOI] [PubMed] [Google Scholar]
- Jacobson JG, Yang K, Baines JD, Homa FL. Linker insertion mutations in the herpes simplex virus type 1 UL28 gene: effects on UL28 interaction with UL15 and UL33 and identification of a second-site mutation in the UL15 gene that suppresses a lethal UL28 mutation. J Virol. 2006;80(24):12312–23. doi: 10.1128/JVI.01766-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kinchington PR, Hougland JK, Arvin AM, Ruyechan WT, Hay J. The Varicella-zoster virus immediate-early protein IE62 is a major component of virus particles. J Virol. 1992;66(1):359–66. doi: 10.1128/jvi.66.1.359-366.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koslowski KM, Shaver PR, Casey JT, 2nd, Wilson T, Yamanaka G, Sheaffer AK, Tenney DJ, Pederson NE. Physical and functional interactions between the herpes simplex virus UL15 and UL28 DNA cleavage and packaging proteins. J Virol. 1999;73(2):1704–7. doi: 10.1128/jvi.73.2.1704-1707.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koslowski KM, Shaver PR, Wang XY, Tenney DJ, Pederson NE. The pseudorabies virus UL28 protein enters the nucleus after coexpression with the herpes simplex virus UL15 protein. J Virol. 1997;71(12):9118–23. doi: 10.1128/jvi.71.12.9118-9123.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krosky PM, Underwood MR, Turk SR, Feng KW, Jain RK, Ptak RG, Westerman AC, Biron KK, Townsend LB, Drach JC. Resistance of human cytomegalovirus to benzimidazole ribonucleosides maps to two open reading frames: UL89 and UL56. J Virol. 1998;72(6):4721–8. doi: 10.1128/jvi.72.6.4721-4728.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lamberti C, Weller SK. The herpes simplex virus type 1 UL6 protein is essential for cleavage and packaging but not for genomic inversion. Virology. 1996;226(2):403–7. doi: 10.1006/viro.1996.0668. [DOI] [PubMed] [Google Scholar]
- Lamberti C, Weller SK. The herpes simplex virus type 1 cleavage/packaging protein, UL32, is involved in efficient localization of capsids to replication compartments. J Virol. 1998;72(3):2463–73. doi: 10.1128/jvi.72.3.2463-2473.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luck S, Sharland M, Griffiths P, Jenkins SM. Advances in the antiviral therapy of herpes virus infection in children. Expert Rev Anti Infect Ther. 2006;4(6):1005–20. doi: 10.1586/14787210.4.6.1005. [DOI] [PubMed] [Google Scholar]
- McNab AR, Desai P, Person S, Roof LL, Thomsen DR, Newcomb WW, Brown JC, Homa FL. The product of the herpes simplex virus type 1 UL25 gene is required for encapsidation but not for cleavage of replicated viral DNA. J Virol. 1998;72(2):1060–70. doi: 10.1128/jvi.72.2.1060-1070.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Newcomb WW, Brown JC. Inhibition of herpes simplex virus replication by WAY-150138: assembly of capsids depleted of the portal and terminase proteins involved in DNA encapsidation. J Virol. 2002;76(19):10084–8. doi: 10.1128/JVI.76.19.10084-10088.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Newcomb WW, Homa FL, Thomsen DR, Brown JC. In vitro assembly of the herpes simplex virus procapsid: formation of small procapsids at reduced scaffolding protein concentration. J Struct Biol. 2001a;133(1):23–31. doi: 10.1006/jsbi.2001.4329. [DOI] [PubMed] [Google Scholar]
- Newcomb WW, Juhas RM, Thomsen DR, Homa FL, Burch AD, Weller SK, Brown JC. The UL6 gene product forms the portal for entry of DNA into the herpes simplex virus capsid. J Virol. 2001b;75(22):10923–32. doi: 10.1128/JVI.75.22.10923-10932.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogasawara M, Suzutani T, Yoshida I, Azuma M. Role of the UL25 gene product in packaging DNA into the herpes simplex virus capsid: location of UL25 product in the capsid and demonstration that it binds DNA. J Virol. 2001;75(3):1427–36. doi: 10.1128/JVI.75.3.1427-1436.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patel AH, MacLean JB. The product of the UL6 gene of herpes simplex virus type 1 is associated with virus capsids. Virology. 1995;206(1):465–78. doi: 10.1016/s0042-6822(95)80062-x. [DOI] [PubMed] [Google Scholar]
- Patel AH, Rixon FJ, Cunningham C, Davison AJ. Isolation and characterization of herpes simplex virus type 1 mutants defective in the UL6 gene. Virology. 1996;217(1):111–23. doi: 10.1006/viro.1996.0098. [DOI] [PubMed] [Google Scholar]
- Proux-Gillardeaux V, Galli T, Callebaut I, Mikhailik A, Calothy G, Marx M. D53 is a novel endosomal SNARE-binding protein that enhances interaction of syntaxin 1 with the synaptobrevin 2 complex in vitro. Biochem J. 2003;370(Pt 1):213–21. doi: 10.1042/BJ20021309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reefschlaeger J, Bender W, Hallenberger S, Weber O, Eckenberg P, Goldmann S, Haerter M, Buerger I, Trappe J, Herrington JA, Haebich D, Ruebsamen-Waigmann H. Novel non-nucleoside inhibitors of cytomegaloviruses (BAY 38-4766): in vitro and in vivo antiviral activity and mechanism of action. J Antimicrob Chemother. 2001;48(6):757–67. doi: 10.1093/jac/48.6.757. [DOI] [PubMed] [Google Scholar]
- Reynolds AE, Fan Y, Baines JD. Characterization of the U(L)33 gene product of herpes simplex virus 1. Virology. 2000;266(2):310–8. doi: 10.1006/viro.1999.0090. [DOI] [PubMed] [Google Scholar]
- Salmon B, Baines JD. Herpes simplex virus DNA cleavage and packaging: association of multiple forms of U(L)15-encoded proteins with B capsids requires at least the U(L)6, U(L)17, and U(L)28 genes. J Virol. 1998;72(4):3045–50. doi: 10.1128/jvi.72.4.3045-3050.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schang LM. Herpes simplex viruses in antiviral drug discovery. Curr Pharm Des. 2006;12(11):1357–70. doi: 10.2174/138161206776361174. [DOI] [PubMed] [Google Scholar]
- Scheffczik H, Savva CG, Holzenburg A, Kolesnikova L, Bogner E. The terminase subunits pUL56 and pUL89 of human cytomegalovirus are DNA-metabolizing proteins with toroidal structure. Nucleic Acids Res. 2002;30(7):1695–703. doi: 10.1093/nar/30.7.1695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scholz B, Rechter S, Drach JC, Townsend LB, Bogner E. Identification of the ATP-binding site in the terminase subunit pUL56 of human cytomegalovirus. Nucleic Acids Res. 2003;31(5):1426–33. doi: 10.1093/nar/gkg229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Severini A, Scraba DG, Tyrrell DL. Branched structures in the intracellular DNA of herpes simplex virus type 1. J Virol. 1996;70(5):3169–75. doi: 10.1128/jvi.70.5.3169-3175.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheaffer AK, Newcomb WW, Gao M, Yu D, Weller SK, Brown JC, Tenney DJ. Herpes simplex virus DNA cleavage and packaging proteins associate with the procapsid prior to its maturation. J Virol. 2001;75(2):687–98. doi: 10.1128/JVI.75.2.687-698.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taus NS, Baines JD. Herpes simplex virus 1 DNA cleavage/packaging: the UL28 gene encodes a minor component of B capsids. Virology. 1998;252(2):443–9. doi: 10.1006/viro.1998.9475. [DOI] [PubMed] [Google Scholar]
- Thoma C, Borst E, Messerle M, Rieger M, Hwang JS, Bogner E. Identification of the interaction domain of the small terminase subunit pUL89 with the large subunit pUL56 of human cytomegalovirus. Biochemistry. 2006;45(29):8855–63. doi: 10.1021/bi0600796. [DOI] [PubMed] [Google Scholar]
- Townsend LB, Devivar RV, Turk SR, Nassiri MR, Drach JC. Design, synthesis, and antiviral activity of certain 2,5,6-trihalo-1-(beta-D-ribofuranosyl)benzimidazoles. J Med Chem. 1995;38(20):4098–105. doi: 10.1021/jm00020a025. [DOI] [PubMed] [Google Scholar]
- Underwood MR, Harvey RJ, Stanat SC, Hemphill ML, Miller T, Drach JC, Townsend LB, Biron KK. Inhibition of human cytomegalovirus DNA maturation by a benzimidazole ribonucleoside is mediated through the UL89 gene product. J Virol. 1998;72(1):717–25. doi: 10.1128/jvi.72.1.717-725.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Zeijl M, Fairhurst J, Jones TR, Vernon SK, Morin J, LaRocque J, Feld B, O'Hara B, Bloom JD, Johann SV. Novel class of thiourea compounds that inhibit herpes simplex virus type 1 DNA cleavage and encapsidation: resistance maps to the UL6 gene. J Virol. 2000;74(19):9054–61. doi: 10.1128/jvi.74.19.9054-9061.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Visalli RJ. Novel compounds for the treatment of varicella-zoster virus infections. Expert Opin Ther Patents. 2004;14(3):355–365. [Google Scholar]
- Visalli RJ, Fairhurst J, Srinivas S, Hu W, Feld B, DiGrandi M, Curran K, Ross A, Bloom JD, van Zeijl M, Jones TR, O'Connell J, Cohen JI. Identification of small molecule compounds that selectively inhibit varicella-zoster virus replication. J Virol. 2003;77(4):2349–58. doi: 10.1128/JVI.77.4.2349-2358.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Visalli RJ, van Zeijl M. DNA encapsidation as a target for anti-herpesvirus drug therapy. Antiviral Res. 2003;59(2):73–87. doi: 10.1016/s0166-3542(03)00108-6. [DOI] [PubMed] [Google Scholar]
- Weber O, Bender W, Eckenberg P, Goldmann S, Haerter M, Hallenberger S, Henninger K, Reefschlager J, Trappe J, Witt-Laido A, Ruebsamen-Waigmann H. Inhibition of murine cytomegalovirus and human cytomegalovirus by a novel non-nucleosidic compound in vivo. Antiviral Res. 2001;49(3):179–89. doi: 10.1016/s0166-3542(01)00127-9. [DOI] [PubMed] [Google Scholar]
- White CA, Stow ND, Patel AH, Hughes M, Preston VG. Herpes simplex virus type 1 portal protein UL6 interacts with the putative terminase subunits UL15 and UL28. J Virol. 2003;77(11):6351–8. doi: 10.1128/JVI.77.11.6351-6358.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wills E, Scholtes L, Baines JD. Herpes simplex virus 1 DNA packaging proteins encoded by UL6, UL15, UL17, UL28, and UL33 are located on the external surface of the viral capsid. J Virol. 2006;80(21):10894–9. doi: 10.1128/JVI.01364-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang K, Baines JD. The putative terminase subunit of herpes simplex virus 1 encoded by UL28 is necessary and sufficient to mediate interaction between pUL15 and pUL33. J Virol. 2006;80(12):5733–9. doi: 10.1128/JVI.00125-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang K, Homa F, Baines JD. Putative terminase subunit of herpes simplex virus 1 form a complex in the cytoplasm and interact with portal protein in the nucleus. J Virol. 2007;81(12):6419–33. doi: 10.1128/JVI.00047-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu D, Weller SK. Genetic analysis of the UL 15 gene locus for the putative terminase of herpes simplex virus type 1. Virology. 1998a;243(1):32–44. doi: 10.1006/viro.1998.9041. [DOI] [PubMed] [Google Scholar]
- Yu D, Weller SK. Herpes simplex virus type 1 cleavage and packaging proteins UL15 and UL28 are associated with B but not C capsids during packaging. J Virol. 1998b;72(9):7428–39. doi: 10.1128/jvi.72.9.7428-7439.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X, Efstathiou S, Simmons A. Identification of novel herpes simplex virus replicative intermediates by field inversion gel electrophoresis: implications for viral DNA amplification strategies. Virology. 1994;202(2):530–9. doi: 10.1006/viro.1994.1375. [DOI] [PubMed] [Google Scholar]


