Figure 1.
Inhibition of Autophagy Leads to Impairment of Proteasomal Degradation
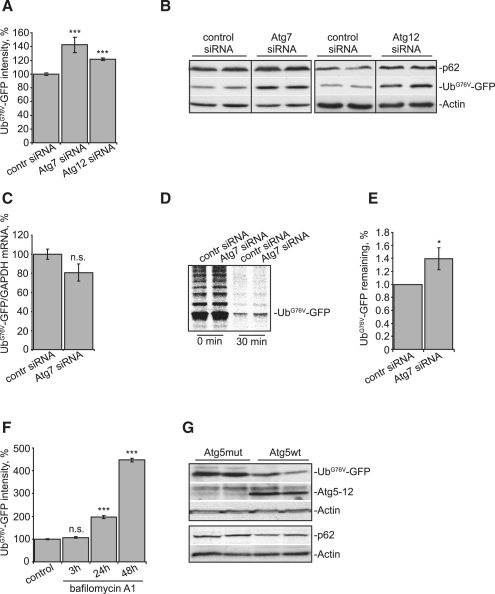
(A and B) siRNA against autophagosomal proteins increases levels of UbG76V-GFP. UbG76V-GFP HeLa cells were transfected with siRNA against two autophagosomal proteins (Atg7 and Atg12), followed by a 72 hr incubation to allow for protein knockdown. GFP fluorescence intensity was quantified by FACS (A), or cells were subjected to immunoblotting (B).
(C) Knockdown of Atg7 does not affect mRNA levels of UbG76V-GFP. mRNA from cells treated as in (A) was used to measure amounts of UbG76V-GFP transcript relative to glyceraldehyde 3-phosphate dehydrogenase (GAPDH) by quantitative PCR.
(D and E) Knockdown of Atg7 slows degradation of UbG76V-GFP. (D) Levels of [35S]methionine UbG76V-GFP were assessed immediately after radioactive pulse (0 min), or following a 30 min chase in the absence of the radiolabel. Bands larger than the main product likely represent different ubiquitinated species. (E) The ratio of [35S]UbG76V-GFP at 30 min to 0 min was significantly higher in atg7 siRNA-treated cells (n = 3). Control values for 30 min/0 min values are normalized to 1, to allow for comparisons of different gels and experiments. In this experiment, the control value at 30 min was 2.27%, whereas that of the Atg7 knockdown was 2.88%. Similar significant findings were obtained in an independent triplicate experiment.
(F) A chemical inhibitor of autophagy, bafilomycin A1, increases levels of UPS reporter in a time-dependent manner. UbG76V-GFP HeLa cells were treated with either DMSO (control) for 48 hr or with 100 nM bafilomycin A1 for the indicated periods of time. GFP fluorescence intensity was quantified by FACS.
(G) Expression of wild-type (wt), but not mutant, Atg5 in atg5−/− MEFs reduces levels of UPS reporter. atg5−/− MEFs were transfected with UbG76V-GFP and either wild-type or mutant (K130R) Atg5 (1:3 ratio). Cells were lysed 48 hr posttransfection and subjected to immunoblotting. Note, only wild-type, but not mutant, Atg5 forms a functional conjugate with Atg12.
For all of the graphs, data are shown as means ± SE for three separate experiments performed in triplicate. ∗p < 0.05, ∗∗∗p < 0.005, t test; all other comparisons are not significant (n.s.).