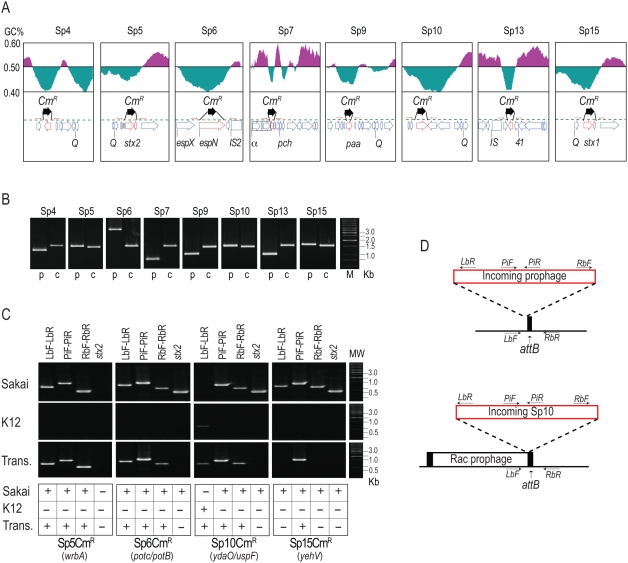
Figure 5. Construction of CmR-marked prophages and analysis of their transfer and lysogenization into K-12 strains.
(A) Strategy to construct CmR-marked prophages. The gene organizations and GC contents of the regions flanking each target gene that was replaced by the CmR gene cassette are shown. (B) Verification of gene replacement using PCR. PCR products obtained from the O157 Sakai parent strain (p) and mutant strains carrying the CmR cassette in each prophage (c) are shown. (C,D) PCR examination of the transfer of CmR-marked prophages from O157 Sakai to K-12. PCR products obtained from O157 Sakai (Sakai) and K-12 (K12) cells and the transductants (Trans) of CmR-marked Sp5, Sp6, Sp10, and Sp15 are shown in (C). Absence of the stx2 gene in all transductants was also confirmed. Integration sites of each prophage in O157 Sakai are indicated in parentheses. Integration patterns of each prophage in K-12 and positions of PCR primers used are shown in (D). The chromosomal attachment site (attB) for Sp10 was occupied by the Rac prophage, and incoming Sp10 was integrated into the K-12 chromosome using the attR site of the Rac prophage. In the Sp15-transductant, the CmR-marked Sp15 was not found at the yehV locus, where Sp15 is integrated in O157 Sakai. Our preliminary result with the Sp15 transductants indicates that the CmR gene on Sp15 was transferred to K-12 by a chimeric phage generated by recombination between the Sp5 and the CmR-marked Sp15 genomes (see text).