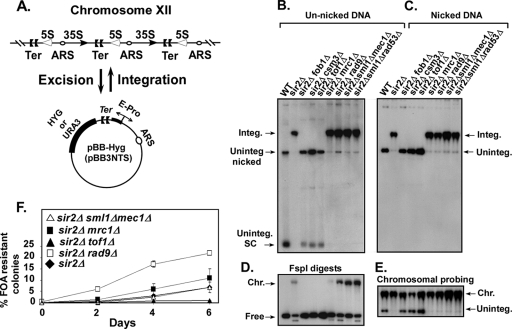
FIG. 1.
Effect of checkpoint genes on plasmid integration into and excision from the Ter sites of rDNA. (A) Schematic representation of the plasmid integration and excision assays; the plasmid pBB3NTS (pBB-Hyg) contained EXP sequence (which includes the Ter sites and the E-pro promoter). (B) Autoradiogram of a Southern blot showing the intracellular distribution of the reporter plasmid pBB-Hyg DNA resolved without nicking the DNA in an 0.5% agarose gel; a labeled plasmid-specific probe was used for detection of the plasmid DNA. integ., integrated; uninteg., unintegrated; SC, supercoiled DNA; WT, wild type. The lanes are self-explanatory. (C) Autoradiogram of a Southern blot showing the DNA samples shown in panel B but after nicking with NB.BsrD1. (D) Same as in panel B except that the DNA samples were digested with FspI; the lanes from left to right are as labeled in panels B and C. (E) Same as in panel C but probed with labeled chromosomal rDNA to identify the location of the integrated plasmid bands. (F) Kinetics of excision of pBB3NTS (URA3) plasmid rDNA from strains containing the integrated form of the plasmid reporter. Effects of individual deletions of the checkpoint genes tof1, mrc1, and rad9 (all present in a sir2Δ background) on loss of URA3 plasmid as a function of time of growth in nonselective medium. The nine lanes in panels D and E correspond exactly to the nine lanes in panels B and C.