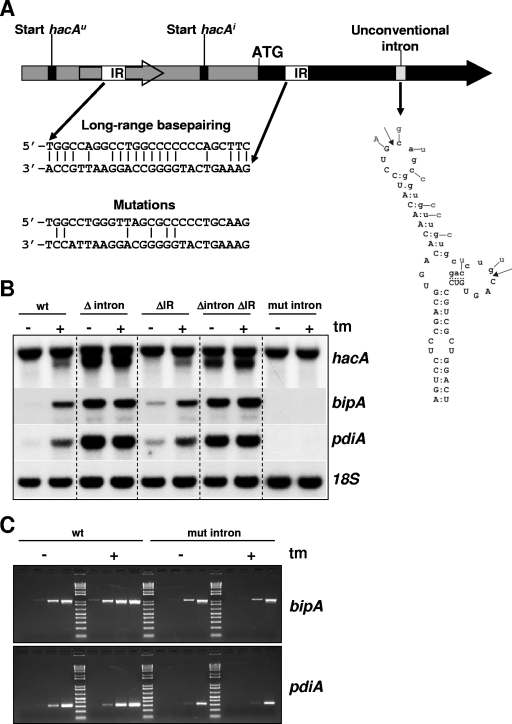
FIG. 5.
Function of the unconventional intron and long-range base pairing in the regulation of the UPR. (A) Schematic overview indicating the unconventional intron and the location of the inverted repeats in relation to other elements present in hacA. The hacA ORF is indicated by a solid black arrow, and the 5′UTR is indicated by a gray box. The inverted repeats (IR) are depicted by open boxes, and the base pairing of the inverted repeats is shown at the bottom. The theoretically remaining base pairings after the introduction of silent mutations are also indicated. The structure of the unconventional intron is depicted by the stem-loop structure, and mutations introduced to prevent splicing by disrupting the structure are indicated. The intron sequence is shown in lowercase type, and cleavage sites are indicated by arrows. (B) Northern blot analysis showing the effect of the different mutations or deletions on the expression and truncation of hacA and on the transcript levels of bipA and pdiA. ER stress was imposed by treatment with tunicamycin (tm), and the gene coding for 18S rRNA was used as a control probe. (C) RT-PCR showing the transcript levels of bipA and pdiA in the wild-type (wt) and mut-intron strains. The four lanes under each condition correspond to 10, 15, 20, and 25 PCR cycles.