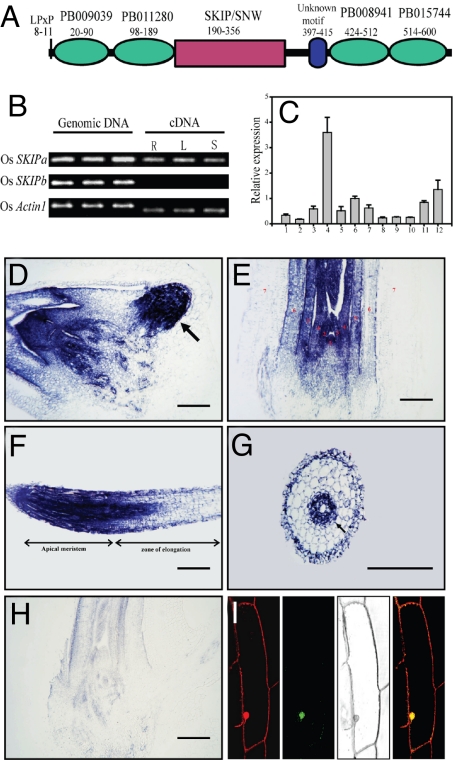
Fig. 1.
Identification of SKIP homologs in rice. (A) Predicted domains and motifs of rice SKIP homolog, OsSKIPa. (B) RT-PCR analysis of OsSKIPb. The genomic DNA and cDNA templates from root (R), young leaf (L), and shoot (S) were used for amplification. Rice Actin1 gene was used as a control. (C) Expression of OsSKIPa in different tissues and organs: 1–4: root, culm, leaf sheath, and leaf lamina, respectively, at heading stage; 5: young panicles (length <1 cm); 6: panicles (length <5 cm); 7: panicles at heading stage; 8: stamen; 9: pistil; 10: etiolated shoot at 1 week after germination; 11: shoot at 1 week after germination; 12: shoot at tillering stage. OsSKIPa transcript in different tissues at 10 days after germination detected by in situ hybridization: radicle (arrow indicates the primordia of lateral root) (D); shoot (shoot apicel meristem labeled with no. 1 and the leaf primordia or mature leaves labeled with nos. 2 to 7 according to the initiation order) (E); root tips (F); cross section of mature zone of root (G); and in situ hybridization control using a sense of OsSKIPa as a probe (H). Bars = 200 μm. (I) Confocal microscopic images of an onion epidermis cell transiently expressing OsSKIPa-GFP fusion protein (Left to Right: propidium iodide stained, GFP, light, merged image).