Summary
Currently, all known translocated effectors of Yersinia are delivered into host cells by type III secretion systems (T3SSs). Pathogenic Yersinia maintain the plasmid-encoded Ysc T3SS for the specific delivery of the well-studied Yop effectors. New horizons for effector biology have opened with the discovery of the Ysps of Y. enterocolitica Biovar 1B, which are translocated into host cells by the chromosome-endoded Ysa T3SS. The reported arsenal of effectors is likely to expand since genomic analysis has revealed gene-clusters in some Yersinia that code for other T3SSs. These efforts also revealed possible type VI secretion (T6S) systems, which may indicate translocation of effectors occurs by multiple mechanisms.
Introduction
The genus Yersinia includes three human pathogens. The most infamous is the black death agent Y. pestis, which causes bubonic plague when it is transmitted by the bite of a flea and pneumonic plague when it is acquired through aerosol transmission [1]. In contrast, Y. enterocolitica and Y. pseudotuberculosis are enteropathogens transmitted by consumption of contaminated food or water [2]. These two organisms cause gastrointestinal syndromes that can develop into fatal septicemia in patients with compromised or underdeveloped immune systems. Regardless of the species and type of disease that ensues, translocation of toxic virulence effectors into host cells by type III secretion (T3S) systems plays an essential role in determining the outcome of a Yersinia infection. There are two recognized groups of effector proteins delivered by T3SSs among the Yersinia; the Yops and the Ysps.
Yop effectors and the innate immune response
Interestingly, despite the various modes of transmission and diseases caused by the pathogenic Yersinia, they commonly have the plasmid-encoded Ysc T3SS for the delivery of six Yop effectors (YopE, YopH, YpkA/YopO, YopM , YopJ/P and YopT) (Table 1). Efforts to determine the biochemical activities of the Yops have revealed many targets are cellular components that influence the host innate immune response (Figure1). This arm of the immune system serves as the front line of defense against an invasive pathogen [3]. Macrophages and neutrophils are important elements of the early innate immune response and serve as sentries that take up and then inactivate invading bacteria. These cells, which act along with other facets of the innate immune response, are induced following the recognition of pathogen-associated molecular patterns (PAMPS) by the family of pattern-recognition receptors (PRRs) located on a variety of cell types. In response to PRR agonists or PAMPS, such as LPS or flagellin, signals are transduced and converged to activate multiple MAPKs and the NF-κB signaling pathways. This inflammatory response in turn leads to the production of numerous proinflammatory cytokines, including TNF-α, IL-12 and IL-18.
Table 1.
Virulence effector proteins of pathogenic Yersinia
| Effectors | T3SS | Gene Locations | Biochemical Functions/Characteristics | Cellular Targets | Cellular Effects |
|---|---|---|---|---|---|
| Pathogenic Y. pestis, Y. psuedotuberculosis and Y. enterocolitica | |||||
| YopE | Ysc/Ysa | Plasmid | Rho GAP mimicry | RhoA, Rac1, Cdc42 | Disruption of actin cytoskeleton Inhibition of phagocytosis |
| YopH | Ysc | Plasmid | Protein tyrosine phosphatase | Focal adhesion complexes (p130Cas, FAK, paxillin, Fyb, SKAP-HOM, Crk, Pyk2) Other proteins LAT, SLP-76, Lck |
Disruption of actin cytoskeleton Disruption of phagocytosis Inhibition of chemoattractant protein (MCP1) production (P13K/Akt signaling) Suppression of adaptive immune response (cytokine IL-2) |
| YpkA/YopO | Ysc | Plasmid | Serine/threonine kinase RhoGDI mimicry Actin binding |
Gαq, RhoA, Rac1 Actin (kinase activity activation) | Disruption of actin cytoskeleton Disruption of phagocytosis Inhibition of Gαq signaling |
| YopM | Ysc | Plasmid | Localization to nucleus Twelve to twenty tandem leucine-rich repeats (~20 amino-acid) |
Rsk1 and Prk2? | Inhibition of cytokine production (IL-15 and IL-15Rα) Depletion of NK cells |
| YopJ/P | Ysc/Ysa | Plasmid | Acetyltransferase | MAPKK and IKK family member proteins | Inhibition of MAPK and NF-κB signaling pathways Suppression of proinflammatory cytokine and chemokine production (TNF-α, IL-8, IL-12, IL-18, etc.) Induction of apoptosis |
| YopT | Ysc | Plasmid | Cysteine protease | RhoA, Rac1, Cdc42 | Disruption of actin cytoskeleton Inhibition of phagocytosis |
| Highly Pathogenicity Y. enterocolitica Biovar 1B | |||||
| YspA | Ysa | Chromosome | ? | ? | ? |
| YspL | Ysa | Chromosome | Two separate 15 or 17 amino-acid repeats | ? | ? |
| YspP | Ysa | Chromosome | Protein tyrosine phosphatase | ? | ? |
| YspF | Ysa | Chromosome | ? | ? | ? |
| YspE | Ysa | Chromosome | ADP-ribosyltransferase? | ? | ? |
| YspI | Ysa | Chromosome | Ca2+ binding protein Three tandem 12 amino-acid repeats |
? | ? |
| YspK | Ysa | Chromosome | Serine/threonine kinase | E2 ubiquitin-conjugating enzymes | Inhibition of NF-κB signaling pathway |
| YspM | Ysa | Chromosome | GDSL lipase motif | ? | Growth inhibition of Saccharomyces cerevisiae |
Figure 1.
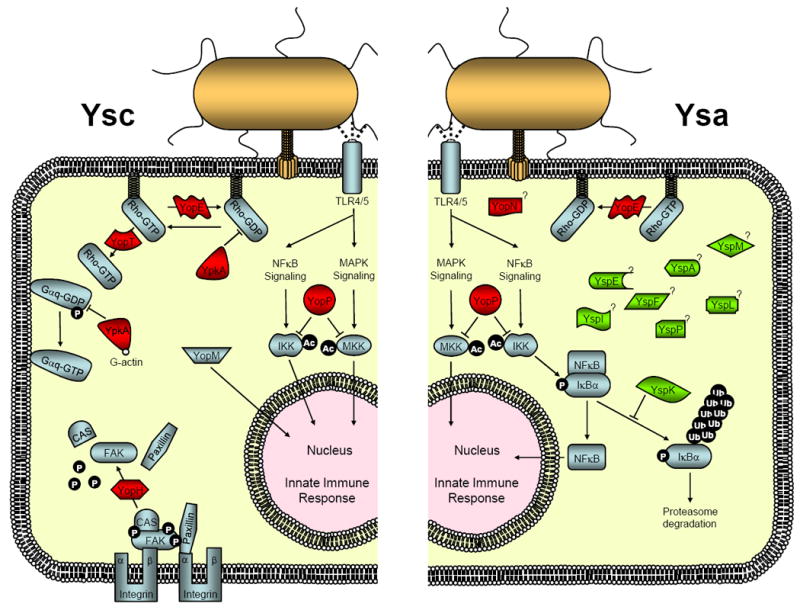
Cellular targets and signaling pathways targeted by Yersinia effectors translocated by the Ysc and Ysa T3SSs. Left side - Effectors delivered by the Ysc T3SS found in Y. pestis, Y. pseudotuberculosis and Y. enterocolitica. Rho family GTPases are targeted by YopE, YpkA/YopO and YopT. The N-terminal serine/threonine kinase domain of YpkA/YopO phosphorylates Gαq to prevent activation of trimeric G proteins. YopH tyrosine phosphatase interacts and dephosphorylates proteins within focal adhesion complexes to inhibit phagocytosis. YopJ/P acetylates serine and threonine residues in the kinase activation loop of MAPKs and IKKs to prevent them from activation through phosphorylation. Inhibition of MAPKs and IKKs limits expression of proinflammatory genes. YopM traffics to the nucleus where it may affect gene transcription networks. Right side - Effectors delivered by the Ysa T3SS of Y. enterocolitica Biovar 1B. Presumably, YopE and YopJ/P function as described above. YopN is injected into cells but it does not have a defined effector function. YspK interacts with E2 ubiquitin-conjugating enzymes and may interfere with proteasome-mediated degradation of IκBα, negative factor controlling the activation of NF-kB thereby inhibiting the expression of proinflammatory genes.
Given the important role that the MAPKs and NF-κB signaling pathways play in the earliest stages of the immune response, it is not surprising that one strategy of pathogenic Yersinia is to interfere with these essential signaling components [4-6]. It is well established that YopJ/P is a potent inhibitor of multiple NF-κB and MAPK pathways [7]. Recent studies have clarified the biochemical nature of YopJ/P activity by revealing it is an enzyme that acetylates serine and threonine residues in the activation loop of MKKs and IKKs [8-10]. This modification effectively blocks the residues from being phosphorylated and results in an inhibition of their activation thereby affecting cytokine production and inducing apoptosis of macrophages.
Phagocytosis of bacteria requires reorganization of the actin cytoskeleton through a process that is regulated by Rho family GTPases [11]. YopE is a Rho GTPase-activating protein (RhoGAP) which has been shown to stimulate GTP hydrolysis, thereby interfering with actin depolymerization mediated by the Rho family of proteins [12,13]. In addition to GTPase activation, it has been shown that membrane anchoring is also required for the activation of RhoA, Rac1 and Cdc42. Membrane anchoring occurs as the result of a post-translational modification resulting in C-terminal prenylation at a cysteine residue. YopT acts as a cysteine protease that removes this lipid modification through cleavage at a site a few residues upstream of the prenylated cysteine [14,15]. The resulting mislocalization inhibits normal signaling function [13,16].
Rho family members are further controlled by Rho guanine nucleotide dissociation inhibitors (RhoGDI). RhoGDI binds and sequesters inactive GDP-bound Rho family members primarily in the cytoplasm. YpkA/YopO contains a C-terminal domain that interacts with RhoA and Rac1 to mimic RhoGDI thereby preventing nucleotide exchange and activation [17]. Interestingly, YpkA/YopO also has an N-terminally located serine/threonine kinase domain that is activated by host cell derived actin, but its substrate remained a mystery for many years [18]. Importantly, a group of G proteins that respond to G protein coupled receptors have emerged as the YpkA/YopO kinase targets [19]. In a recent elegant study, it was demonstrated that YpkA/YopO specifically phosphorylates Ser47 of Gαq, a key residue located in the highly conserved diphosphate binding loop of the GTPase fold [19]. This phosphorylation impairs nucleotide binding and prevents Gαq-mediated cellular processes, including the subsequent activation of RhoA which would be expected to limit bacterial phagocytosis by cells. However, the exact role of YpkA/YopO kinase in affecting cellular activities remains an avenue open for further investigation since there are a myriad of signaling pathways affected by this Gαq group of G protein signaling molecules.
Phagocytosis is additionally inhibited by YopH, a protein tyrosine phosphatase that antagonizes several signaling pathways [20-22]. YopH specifically localizes to focal adhesion complexes where it interacts with and dephosphorylates proteins such as FAK, Cas and paxillin [21,23]. These proteins are involved in β1 integrin mediated phagocytic events brought about by the surface protein invasin of Y. enterocolitica and Y. pseudotuberculosis during epithelial cell invasion. Likewise, in macrophages, where uptake events can be mediated by complement receptors or Fc receptors, YopH dephosphorylates focal adhesin complex-associated proteins, including Cas, SKAP-HOM, Fyb and the FAK-homolog Pyk2 [24,25].
The remaining effector targeted by the Ysc T3SS is YopM, which has no clearly established function. YopM is known to traffic to the nucleus when introduced into HeLa cells. One study of the role that YopM plays during Y. pestis infection provides intriguing clues for future efforts. YopM function correlates with the global depletion of natural killer cells by affecting expression of IL-15 and IL-15Rα [26].
Ysp effectors of Y. enterocolitica Biovar 1B
While the Ysc T3SS is important for Yersinia virulence, it is now clear that some isolates of Yersinia utilize additional T3SSs to deliver virulence effectors into targeted host cells. The highly pathogenic Y. enterocolitica Biovar 1B carries the Ysa pathogenicity island (Ysa-PI) encoding a T3SS which is distinctly different from the Ysc T3SS and is more related to the Mxi-Spa T3SS of Shigella species [27-29]. The Ysa-PI is part of a larger region of the chromosome, called the plasticity zone, containing numerous other genes implicated in virulence [30].
Studies using the mouse model of Yersiniosis demonstrated that the Ysa T3SS plays an important role in Y. enterocolitica colonization of gastrointestinal tissues during the earliest stages of an infection [31]. This observation brings new attention to how enteropathogenic Yersinia colonizes the intestine and overcomes immune barriers presented by the host at this location. It may be that the Ysc T3SS is important for systemic stages of infection, while the Ysa T3SS is selective for gastrointestinal infection. Evaluating this model is dependent upon defining and delineating the function of the virulence effectors delivered by the Ysa T3SS.
The first proteins determined to be Ysps effector by virtue of their being substrates exported by the Ysa T3SS unexpectedly turned out to be YopE, YopN and YopJ/P, three proteins also exported by the Ysc T3SS [32] [27] (Table 1). As described above, YopE and YopJ/P are known to be effectors, but it is possible that YopN may also have an unrecognized effector function. It was further demonstrated that Y. enterocolitica Biovar 1B delivers YopJ/P into cultured macrophages and suppresses production of TNF-α by utilizing either the Ysa or Ysc pathways [32]. However, gaining a comprehensive understanding of how the Ysa T3SS mechanistically influences pathogenesis will depend on efforts to distinguish the functions of the effectors it delivers.
In this regard, eight additional translocated effectors have been identified (YspA, YspE, YspF, YspI, YspK, YspL, YspM and YspP) (Table 1). YspA was recognized from analysis of genes within the Ysa-PI [27]. Recently, a comprehensive proteomics approach defined the complexity of the Ysa T3SS secretome [33]. This effort confirmed YspA as an effector and additionally identified YspE, YspF, YspI, YspK, YspL and YspP. Each of these effectors was shown to be necessary for full virulence of Y. enterocolitica Biovar 1B in the mouse gastroenteritis competitive index assay [33]. Finally, YspM was identified later as an effector produced by a subset of Biovar 1B strains. When including YopE, YopN and YopJ/P, the Ysa T3SS appears to deliver a collection of ten, and potentially eleven, effectors depending upon the strain examined.
It is striking that the genes encoding the effectors delivered by the Ysa T3SS are dispersed throughout the genome [33] (Figure 2). One ysp is located within the Ysa-PI, three are located on plasmid pYV, but the seven remaining map to sites dispersed throughout the chromosome. The lack of ysp gene co-localization inspires the idea that the evolution of Y. enterocolitica Biovar 1B involved numerous genetic lateral transfer events. The dispersed gene distribution combined with the complexity of the Ysps suggests that Y. enterocolitica Biovar 1B has experienced strong selective pressure to maintain the Ysa T3SS. This may reflect how this group of Yersina has adapted to survive differently in the gastrointestinal environment and may help to explain the severity of human infections they cause.
Figure 2.
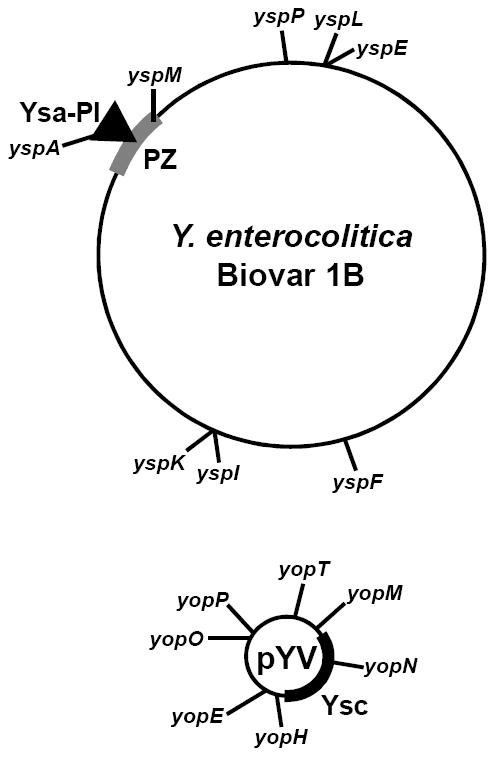
Genes of Y. enterocolitica Biovar 1B that encode effectors translocated into host cells by the Ysa T3SS are dispersed in the genome. Depicted is the chromosome with labels indicating the relative locations of effector-encoding ysp genes and the location of the YSA-PI encoding the Ysa T3SS. The Ysa-PI is within a region of the chromosome rich with other genes implicated in virulence called the plasticity zone (PZ). Plasmid pYV carries genes encoding the Ysc T3SS (Ysc) and the effector Yops. A similar plasmid is present in Y. pestis and Y. pseudotuberculosis.
With the exception of YopE and YopJ/P, the cellular targets of the effector proteins delivered by the Ysa T3SS are largely open for investigation (Table 1, and Figure1). Several of the Ysps contain regions or domains that can be predicted to have particular functions. YspK serine/threonine kinase activity and YspP tyrosine phosphatase activity were demonstrated using in vitro-based biochemical assays [33]. Furthermore, YspK shares 91% identity with OspG, an effector produced by Shigella flexneri [34]. OspG can bind ubiquitinylated E2 conjugating enzymes, including UbcH5b that is responsible for targeting the degradation of many important signaling factors such as phospho-IκBα by the proteasome [34]. By interfering with phospho-IκBα degradation, its complex with NF-κB is maintained, which suppresses activation of this proinflammatory pathway. Using yeast two-hybrid analysis, YspK has also been shown to interact with E2 proteins (Matsumoto and Young, unpublished data). This result suggests YspK and OspG have similar functions in targeting cellular components to limit the innate immune response (Figure 1). YspM is predicted to be a GDSL lipase and when expressed in Saccharomyces cerevisiae it prevents cell growth. It is tantalizing to consider the predicted lipase activity may be targeting an essential cellular component but this remains to be demonstrated [35]. Other Ysps with interesting domains predicted by computational analysis include YspE with an ADP-ribosyltransferase motif (Matsumoto and Young, unpublished results). In other pathogenic bacteria ADP-ribosyltransferases are among the most potent virulence factors, marking YspE as a high priority for exploration of its cellular target [36]. Likewise, a calcium-binding motif predicted for YspI is a unique characteristic of an effector. This may indicate YspI belongs to a new class of virulence factors affecting cellular activities through a novel mechanism trigged by calcium sensing. Clearly these are speculative predictions, but they are presented to highlight the potential within the study of the Ysp effector proteins to reveal new facets of host-pathogen interactions.
New Yersinia effectors yet to be discovered
Genomic sequencing of numerous strains of Y. pestis and Y. pseudotuberculosis have revealed a locus with the potential to encode a T3SS that is different than either the Ysc or Ysa T3SSs [37,38]. This system resembles the Ssa T3SS of Salmonella enterica [39]. It has also been reported that this locus is present in some Y. enterocolitica serotype O:3 strains [40]. To date, no clear link between this T3SS and virulence has been established. On another front, one controversial study has suggested the insecticidal toxin-like proteins (IT-like proteins) of Y. pestis and Y. pseudotuberculosis may form a new group of effectors [41]. This assertion was based on the observation that in vitro ectopic expression of these IT-like proteins led to secretion, and in one case even translocation into insect and mammalian cells, by the Ysc T3SS. The idea that the Ysc T3S is the native secretion pathway for the insecticidal toxin has recently been challenged, but it remains a formal possibility that they are exported by the chromosomal-encoded Ssa-like T3SS [42].
Do Yersinia translocate effectors by other mechanisms?
Direct translocation of effectors into host cells by Gram-negative bacteria can additionally involve T4SSs and T6SSs. While no T4SS has been described, recent genomic analysis has suggested the presence of several potential T6SSs encoded by loci dispersed among Y. pestis and Y. pseudotuberculosis genomes [43,44]. Some of these gene-clusters could encode proteins homologous to the Hcp- and VgrG-families of secreted proteins and effectors. With the recent recognition that T6SS translocate effectors into host cells there appears to be abundant opportunity for new effectors to be discovered.
Conclusions
The Yop effectors are nearly identical between Y. pestis, Y. pseudotuberculosis and Y. enterocolitica. The study of pathogenic Yersinia presents an opportunity to examine how these effectors are utilized by pathogenic species causing different types of infections. The discovery of the Ysa T3SS of Y. enterocolitica Biovar 1B expands on this theme of comparative pathology by further revealing that some effectors, like YopE and YopJ/P, can contribute to disease when delivered through other pathways, such as the Ysa T3SS. However, the new twists on effector biology will come from further pursuing the functions of newly discovered effectors such as the Ysps. With the recognition of possible translocated effectors by T6SSs, there appears to be abundant opportunity for new effectors to be discovered.
Acknowledgments
The authors apologize to colleagues whose work could not be cited due to space limitations. We express our appreciation for the editorial advise of Briana Young. Work in GMY’s laboratory is sponsored by grants from the National Institutes of Health, R21 AI165042 and R21 AI067676.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References and recommended reading
Papers of particular interest, published within the annual period of the review, have been highlighted as:
* of special interest
* * of outstanding interest
- 1.Stenseth NC, Atshabar BB, Begon M, Belmain SR, Bertherat E, Carniel E, Gage KL, Leirs H, Rahalison L. Plague: past, present, and future. PLoS Med. 2008;5:e3. doi: 10.1371/journal.pmed.0050003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bottone EJ. Yersinia enterocolitica: overview and epidemiologic correlates. Microbes Infect. 1999;1:323–333. doi: 10.1016/s1286-4579(99)80028-8. [DOI] [PubMed] [Google Scholar]
- 3.Medzhitov R. Recognition of microorganisms and activation of the immune response. Nature. 2007;449:819–826. doi: 10.1038/nature06246. [DOI] [PubMed] [Google Scholar]
- 4.Shao F. Biochemical functions of Yersinia type III effectors. Curr Opin Microbiol. 2008;11:21–29. doi: 10.1016/j.mib.2008.01.005. [DOI] [PubMed] [Google Scholar]
- 5.Viboud GI, Bliska JB. Yersinia outer proteins: role in modulation of host cell signaling responses and pathogenesis. Annu Rev Microbiol. 2005;59:69–89. doi: 10.1146/annurev.micro.59.030804.121320. [DOI] [PubMed] [Google Scholar]
- 6.Trosky JE, Liverman AD, Orth K. Yersinia outer proteins: Yops. Cell Microbiol. 2008;10:557–565. doi: 10.1111/j.1462-5822.2007.01109.x. [DOI] [PubMed] [Google Scholar]
- 7.Orth K. Function of Yersinia effector YopJ. Current Opinion in Microbiology. 2002;5:38–43. doi: 10.1016/s1369-5274(02)00283-7. [DOI] [PubMed] [Google Scholar]
- **8.Mukherjee S, Keitany G, Li Y, Wang Y, Ball HL, Goldsmith EJ, Orth K. Yersinia YopJ acetylates and inhibits kinase activation by blocking phosphorylation. Science. 2006;312:1211–1214. doi: 10.1126/science.1126867. [DOI] [PubMed] [Google Scholar]
- **9.Mittal R, Peak-Chew SY, McMahon HT. Acetylation of MEK2 and I kappa B kinase (IKK) activation loop residues by YopJ inhibits signaling. Proc Natl Acad Sci U S A. 2006;103:18574–18579. doi: 10.1073/pnas.0608995103.. These two studies convincingly demonstrated that acetyltransferase activity of YopJ/P which sheds light on a new biochemical modification that affects cellular processes. This biochemical modification may have broader implications on understanding how mammalian sigal transductions pathways are regulated.
- *10.Hao YH, Wang Y, Burdette D, Mukherjee S, Keitany G, Goldsmith E, Orth K. Structural requirements for Yersinia YopJ inhibition of MAP kinase pathways. PLoS ONE. 2008;3:e1375. doi: 10.1371/journal.pone.0001375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Barbieri JT, Riese MJ, Aktories K. Bacterial toxins that modify the actin cytoskeleton. Annu Rev Cell Dev Biol. 2002;18:315–344. doi: 10.1146/annurev.cellbio.18.012502.134748. [DOI] [PubMed] [Google Scholar]
- 12.Aepfelbacher M. Modulation of Rho GTPases by type III secretion system translocated effectors of Yersinia. Rev Physiol Biochem Pharmacol. 2004;152:65–77. doi: 10.1007/s10254-004-0035-3. [DOI] [PubMed] [Google Scholar]
- *13.Viboud GI, Mejia E, Bliska JB. Comparison of YopE and YopT activities in counteracting host signalling responses to Yersinia pseudotuberculosis infection. Cell Microbiol. 2006;8:1504–1515. doi: 10.1111/j.1462-5822.2006.00729.x. [DOI] [PubMed] [Google Scholar]
- 14.Sorg I, Hoffmann C, Dumbach J, Aktories K, Schmidt G. The C terminus of YopT is crucial for activity and the N terminus is crucial for substrate binding. Infect Immun. 2003;71:4623–4632. doi: 10.1128/IAI.71.8.4623-4632.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Shao F, Merritt PM, Bao Z, Innes RW, Dixon JE. A Yersinia effector and a Pseudomonas avirulence protein define a family of cysteine proteases functioning in bacterial pathogenesis. Cell. 2002;109:575–588. doi: 10.1016/s0092-8674(02)00766-3. [DOI] [PubMed] [Google Scholar]
- 16.Aepfelbacher M, Trasak C, Wilharm G, Wiedemann A, Trulzsch K, Krauss K, Gierschik P, Heesemann J. Characterization of YopT effects on Rho GTPases in Yersinia enterocolitica-infected cells. J Biol Chem. 2003;278:33217–33223. doi: 10.1074/jbc.M303349200. [DOI] [PubMed] [Google Scholar]
- **17.Prehna G, Ivanov MI, Bliska JB, Stebbins CE. Yersinia virulence depends on mimicry of host Rho-family nucleotide dissociation inhibitors. Cell. 2006;126:869–880. doi: 10.1016/j.cell.2006.06.056.. These authors utilized structural analysis in combination with biochemical approaches to demonstrate the C-terminus of YpkA/YopO binds RhoA and Rac1 in manner that mimics RhoGDI. This provides a mechanistic model that supports previous observations of kinase-independent functions for YpkA/YopO.
- 18.Galyov EE, Hakansson S, Forsberg A, Wolf-Watz H. A secreted protein kinase of Yersinia pseudotuberculosis is an indispensable virulence determinant. Nature. 1993;361:730–732. doi: 10.1038/361730a0. [DOI] [PubMed] [Google Scholar]
- **19.Navarro L, Koller A, Nordfelth R, Wolf-Watz H, Taylor S, Dixon JE. Identification of a molecular target for the Yersinia protein kinase A. Mol Cell. 2007;26:465–477. doi: 10.1016/j.molcel.2007.04.025.. This kinase substrate for YpkA/YopO eluded investigators for many years. This elegant study revealed Gαq to be phosphrylated by YpkA at a residue in the diphosphate binding loop that is necessary for nucleotide exchange. This implicates trimeric G proteins in controlling key sigaling pathways that affect the outcome of an infection.
- 20.Persson C, Carbelleira N, Wolf-Watz H, Fallman M. The PTPase YopH inhibits uptake of Yersinia, tyrosine phosphorylation of p130cas and FAK, and the associated accumulation of the proteins in peripheral focal adhesion. EMBO J. 1997;16:2307–2318. doi: 10.1093/emboj/16.9.2307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Persson C, Nordfelth R, Andersson K, Forsberg A, Wolf-Watz H, Fallman M. Localization of the Yersinia PTPase to focal complexes is an important virulence mechanism. Mol Microbiol. 1999;33:828–838. doi: 10.1046/j.1365-2958.1999.01529.x. [DOI] [PubMed] [Google Scholar]
- 22.Black DS, Bliska JB. Identification of p130Cas as a substrate of Yersinia YopH (Yop51), a bacterial protein tyrosine phosphatase that translocates into mammalian cells and targets focal adhesions. Embo J. 1997;16:2730–2744. doi: 10.1093/emboj/16.10.2730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Black DS, Montagna LG, Zitsmann S, Bliska JB. Identification of an amino-terminal substrate-binding domain in the Yersinia tyrosine phosphatase that is required for efficient recognition of focal adhesion targets. Mol Microbiol. 1998;29:1263–1274. doi: 10.1046/j.1365-2958.1998.01014.x. [DOI] [PubMed] [Google Scholar]
- 24.Fallman M, Andersson K, Hakansson S, Magnusson KE, Stendahl O, Wolf-Watz H. Yersinia pseudotuberculosis inhibits Fc receptor-mediated phagocytosis in J774 cells. Infect Immun. 1995;63:3117–3124. doi: 10.1128/iai.63.8.3117-3124.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ruckdeschel K, Roggenkamp A, Schubert S, Heesemann J. Differential contribution of Yersinia enterocolitica virulence factors to evasion of microbicidal action of neutrophils. Infect Immun. 1996;64:724–733. doi: 10.1128/iai.64.3.724-733.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kerschen EJ, Cohen DA, Kaplan AM, Straley SC. The plague virulence protein YopM targets the innate immune response by causing a global depletion of NK cells. Infect Immun. 2004;72:4589–4602. doi: 10.1128/IAI.72.8.4589-4602.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Foultier B, Troisfontaines P, Vertommen D, M M-N, Rider M, Persot C, Cornelis GR. Identification of substrates and chaperone from the Yersinia enterocolitica 1B Ysa type III secretion system. Infect Immun. 2003;71:242–253. doi: 10.1128/IAI.71.1.242-253.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Haller JC, Carlson S, Pederson KJ, Pierson DE. A chromosomally encoded type III secretion pathway in Yersinia enterocolitica is important in virulence. Mol Microbiol. 2000;36:1436–1446. doi: 10.1046/j.1365-2958.2000.01964.x. [DOI] [PubMed] [Google Scholar]
- 29.Young GM. The Ysa type 3 secretion system of Yersinia enterocolitica biovar 1B. Adv Exp Med Biol. 2007;603:286–297. doi: 10.1007/978-0-387-72124-8_26. [DOI] [PubMed] [Google Scholar]
- *30.Thomson NR, Howard S, Wren BW, Holden MT, Crossman L, Challis GL, Churcher C, Mungall K, Brooks K, Chillingworth T, et al. The complete genome sequence and comparative genome analysis of the high pathogenicity Yersinia enterocolitica strain 8081. PLoS Genet. 2006;2:e206. doi: 10.1371/journal.pgen.0020206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Venecia K, Young GM. Environmental regulation and virulence attributes of the Ysa type III secretion system of Yersinia enterocolitica biovar 1B. Infect Immun. 2005;73:5961–5977. doi: 10.1128/IAI.73.9.5961-5977.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Young BM, Young GM. Evidence for targeting of Yop effectors by the chromosomally encoded Ysa type III secretion system of Yersinia enterocolitica. J Bacteriol. 2002;184:5563–5571. doi: 10.1128/JB.184.20.5563-5571.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- **33.Matsumoto H, Young GM. Proteomic and functional analysis of the suite of Ysp proteins exported by the Ysa type III secretion system of Yersinia enterocolitica Biovar 1B. Mol Microbiol. 2006;59:689–706. doi: 10.1111/j.1365-2958.2005.04973.x.. The study revealed a completely new set of Yersinia effectors that affect the outcome of a gastrointestinal infection. This information helped to clarify that the Ysa T3S system was needed for infection of a mammalian host.
- **34.Kim DW, Lenzen G, Page AL, Legrain P, Sansonetti PJ, Parsot C. The Shigella flexneri effector OspG interferes with innate immune responses by targeting ubiquitin-conjugating enzymes. Proc Natl Acad Sci U S A. 2005;102:14046–14051. doi: 10.1073/pnas.0504466102.. This study has important implications on understanding the function of YspK, which is 91% identical to OspG. A series of elegant biochemical approaches and the use of a mouse model of infection revealed OspG interacts with ubiquitin-cojugating enzymes to direct suppression of the proinflammatory pathways and affect disease pathology.
- *35.Witowski SE, Walker KA, Miller VL. YspM, a newly identified Ysa type III secreted protein of Yersinia enterocolitica. J Bacteriol. 2008;190:7315–7325. doi: 10.1128/JB.00861-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Holbourn KP, Shone CC, Acharya KR. A family of killer toxins. Exploring the mechanism of ADP-ribosylating toxins. FEBS J. 2006;273:4579–4593. doi: 10.1111/j.1742-4658.2006.05442.x. [DOI] [PubMed] [Google Scholar]
- 37.Chain PS, Carniel E, Larimer FW, Lamerdin J, Stoutland PO, Regala WM, Georgescu AM, Vergez LM, Land ML, Motin VL, et al. Insights into the evolution of Yersinia pestis through whole-genome comparison with Yersinia pseudotuberculosis. Proc Natl Acad Sci U S A. 2004;101:13826–13831. doi: 10.1073/pnas.0404012101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Parkhill J, Wren BW, Thomson NR, Titball RW, Holden MT, Prentice MB, Sebaihia M, James KD, Churcher C, Mungall KL, et al. Genome sequence of Yersinia pestis, the causative agent of plague. Nature. 2001;413:523–527. doi: 10.1038/35097083. [DOI] [PubMed] [Google Scholar]
- 39.Troisfontaines P, Cornelis GR. Type III secretion: more systems than you think. Physiology (Bethesda) 2005;20:326–339. doi: 10.1152/physiol.00011.2005. [DOI] [PubMed] [Google Scholar]
- 40.Golubov A, Heesemann J, Rakin A. Uncovering genomic differences in human pathogenic Yersinia enterocolitica. FEMS Immunol Med Microbiol. 2003;38:107111. doi: 10.1016/S0928-8244(03)00182-2. [DOI] [PubMed] [Google Scholar]
- 41.Gendlina I, Held KG, Bartra SS, Gallis BM, Doneanu CE, Goodlett DR, Plano GV, Collins CM. Identification and type III-dependent secretion of the Yersinia pestis insecticidal-like proteins. Mol Microbiol. 2007;64:1214–1227. doi: 10.1111/j.1365-2958.2007.05729.x. [DOI] [PubMed] [Google Scholar]
- *42.Hares MC, Hinchliffe SJ, Strong PC, Eleftherianos I, Dowling AJ, Ffrench-Constant RH, Waterfield N. The Yersinia pseudotuberculosis and Yersinia pestis toxin complex is active against cultured mammalian cells. Microbiology. 2008;154:3503–3517. doi: 10.1099/mic.0.2008/018440-0. [DOI] [PubMed] [Google Scholar]
- 43.Bingle LE, Bailey CM, Pallen MJ. Type VI secretion: a beginner’s guide. Curr Opin Microbiol. 2008;11:3–8. doi: 10.1016/j.mib.2008.01.006. [DOI] [PubMed] [Google Scholar]
- *44.Yen YT, Bhattacharya M, Stathopoulos C. Genome-wide in silico mapping of the secretome in pathogenic Yersinia pestis KIM. FEMS Microbiol Lett. 2008;279:56–63. doi: 10.1111/j.1574-6968.2007.01008.x. [DOI] [PubMed] [Google Scholar]


