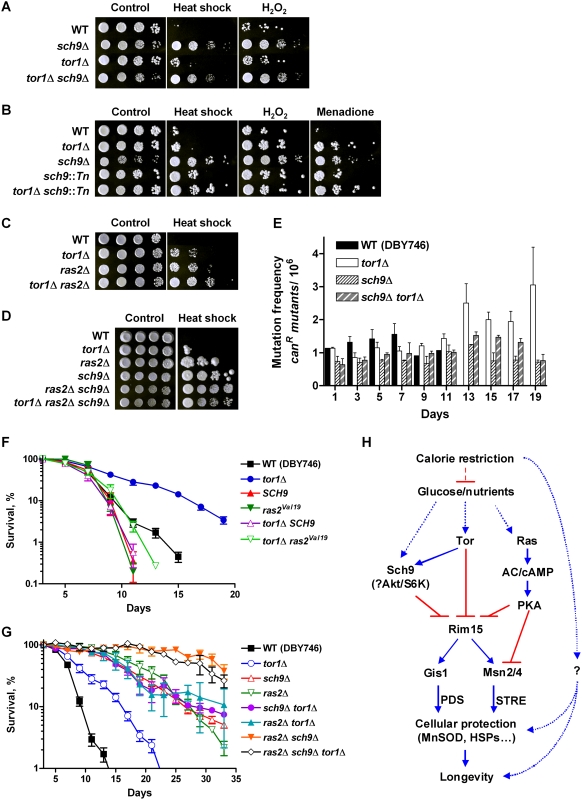
Figure 1. Genetic interactions between Sch9, Tor1, and Ras2 in regulating stress resistance and life span.
(A–D) Day 3 wild type (DBY746) and cells lacking Tor1, Sch9, or Ras2 were challenged with heat shock (55°C: A, 105 min; B, 75 min; C, 150 min; and D, 120 min) or oxidative stresses (H2O2, 100 mM for 60 min; or menadione, 250 µM for 30 min). (E) Mutation frequency over time measured as Canr mutants per million cells. The average of four experiments is shown. Error bars represent SEM. (F) Chronological survival in minimal complete medium (SDC) of wild type, tor1Δ, and mutants overexpressing either SCH9 or constitutively active Ras2 (ras2Val19). (G) Chronological survival of wild type and mutants lacking Tor1, Sch9, Ras2 or combinations. The data represent average of at least 4 experiments. Error bars show SEM. For mean life span calculated from non-linear curve fitting see Table S1. (H) Longevity regulatory pathways in yeast. The nutrient sensing pathways controlled by Sch9, Tor, and Ras converge on the protein kinase Rim15. In turn, the stress response transcription factors Msn2, Msn4, and Gis1 transactivate stress response genes and enhance cellular protection, which lead to life span extension. Pro-longevity effects of CR are partially mediated by Sch9, Tor, and Ras, and may also require additional yet-to-be indentified mechanism(s).