FIGURE 4. RNA interference-mediated silencing of NRF-1 suppresses mRNAs in all 10 nuclear-encoded COX subunit genes and another six nuclear genes important in mitochondrial biogenesis.
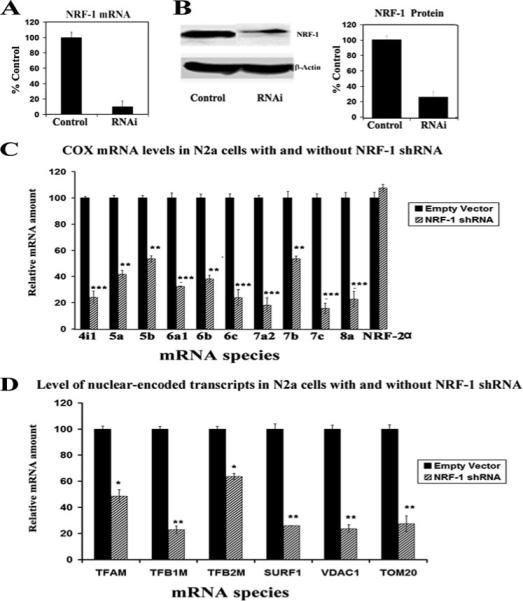
A, N2a cells transfected with NRF-1 shRNA expressed significantly less NRF-1 mRNA than those with empty vectors. B, Western blot reveals a down-regulation of NRF-1 protein in shRNA-transfected neurons. β-Actin served as a loading control. C, N2a cells were transfected with shRNA against NRF-1 (hatched bars) or with empty vectors (solid bars). NRF-2α served as a negative control. All ten COX subunit mRNAs show significant decreases in shRNA-treated samples as compared with those with empty vectors, whereas NRF-2α mRNA remained unchanged. n = 5−6 for each data point; *, p < 0.05; **, p < 0.01; ***, p < 0.001. D, mRNAs were quantified for six other known target genes of NRF-1: TFAM, TFB1M, TFB2M, SURF1, VDAC1, and TOM20 in N2a cells. All six genes showed significant reductions in mRNA expression in NRF-1 shRNA-transfected cells (hatched bars) as compared with empty vector controls (solid bars). n = 5−6 for each data point; *, p < 0.05; **, p < 0.01; ***, p < 0.001.
