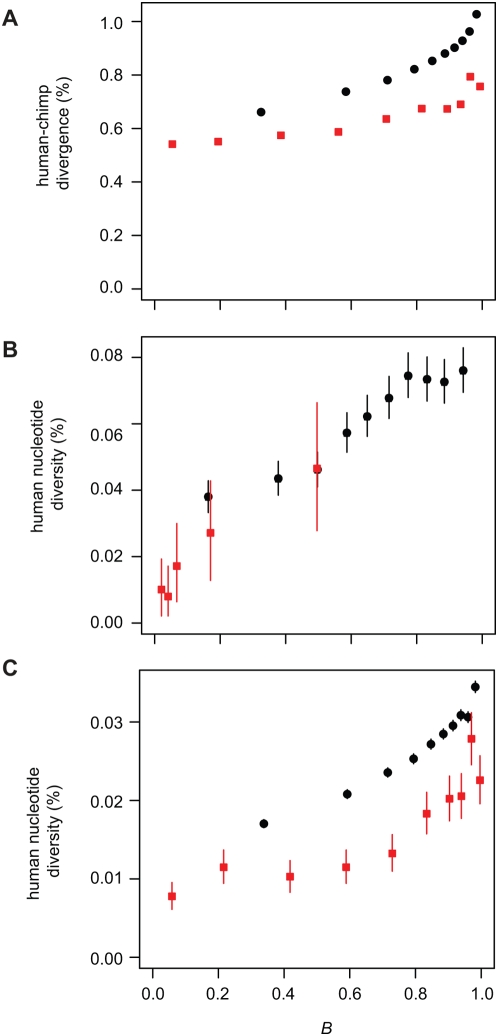
Figure 6. Whole-genome neutral divergence and diversity show strong dependence on the estimated strength of background selection.
(A) Human/chimpanzee divergence from whole-genome alignments for autosomes (black circles) and chromosome X (red squares) versus B (the portion of neutral diversity expected to remain after accounting for background selection). (B) Human nucleotide diversity from Seattle SNPs PGA/EGP [20] data versus B. (C) Human nucleotide diversity from Perlegen [19] data. Estimated diversity is much lower in the Perlegen dataset because it subsamples common variants [19]. Vertical lines are 95% confidence intervals (not visible in (A) because they are smaller than the plotting symbols). Note that although human diversity shows a clear linear relationship to B, a fitted line would not pass through the origin as it should if the 5-species estimates are applicable to recent human evolution. This likely reflects the sharp decrease in human effective population size relative to ancestral primate populations, which is expected to reduce the efficiency of selection on weakly deleterious mutations due to increased genetic drift [31].