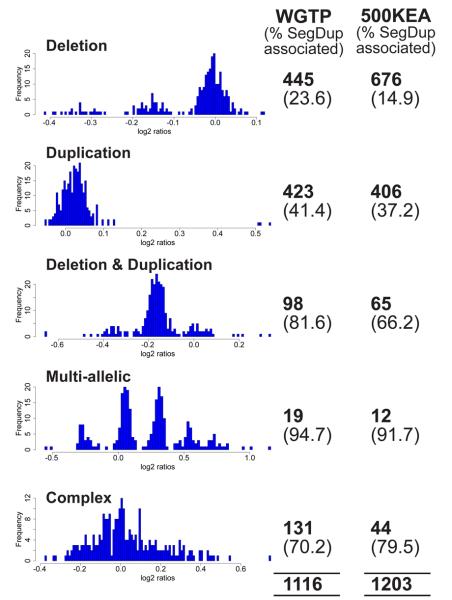
Figure 5. Classes of copy number variants.
CNVs identified from WGTP and 500K EA platforms can be classified from the population distribution of log2 ratios (exemplified with WGTP data) into five different types (see text). Biallelic CNVs (deletions and duplications) can be genotyped if the clusters representing different genotypes are sufficiently distinct. The numbers of each class of CNV identified on WGTP and 500K EA platforms are given, along with the proportion of those CNVs that overlap segmental duplications. The overall proportion of CNVRs overlapping segmental duplications was 20% and 34% on the 500K EA and WGTP platforms, respectively.