Fig. 1.
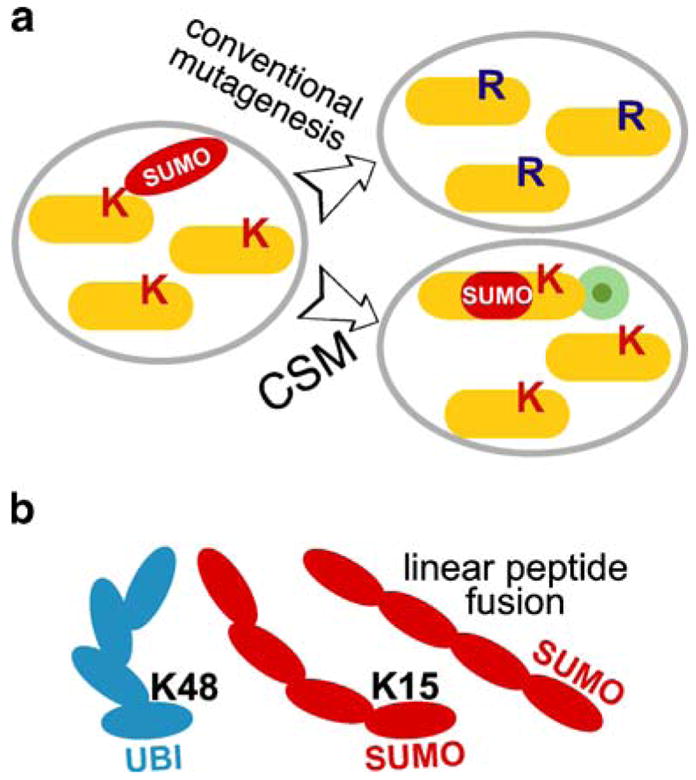
The experimental approach to modeling polysumoylation in vivo. a The advantage of constitutive SUMO modification (CSM) technique over traditional acceptor-site lysine mutagenesis. Ectopic expression of the tagged CSM target (lower right) represents a much better approximation of the wild type situation (left), compared to traditional mutagenesis of the sumoylated sites lysines (K to R; top right). CSM has an added benefit of biochemical/cytological tracking of the CSM pool of a given protein with an epitope tag (green circle). b Tandem peptide fusion of SUMO molecules is a good model of natural polysumoylation because of the proximal NH3-terminal position of the branching site lysine (K15) in SUMO. Branching in a polyubiquitin chain (K48) is shown for comparison
