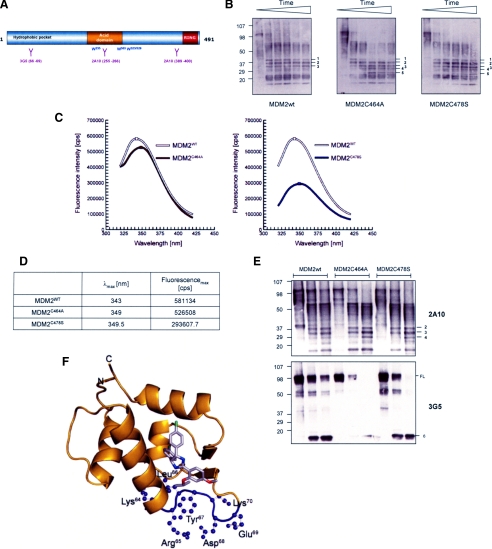
FIGURE 6.
Conformational differences between wt and RING domain mutants of MDM2. A, a schematic representation of MDM2 showing the epitopes for 3G5 and 2A10; the position of the 4 tryptophan residues in MDM2 is marked. B, immunoblot showing wt or mutant forms of MDM2 following limited proteolysis with trypsin. The blot was developed using a mixture of the MDM2 monoclonal antibodies 2A10, 4B2, 3G5, and SMP14. The numbers are used to label the banding pattern; time points used were 0, 5, 10, 15, 20, and 30 min. The data are representative of three separate experiments. C, resolution of fluorescence emission spectra of MDM2wt (white) and the MDM2C464A (brown) and MDM2C478S (blue) RING mutants. The spectra were corrected for associated buffer background signals. D, maxima with corresponding wavelength of MDM2 fluorescence emission spectra. The values were calculated with mathematical n-polynomial based algorithm, where R2 ≥ 0.999. E, a sin B except the time course was 0, 5, and 20 min. The blots were developed using 2A10 (upper panel) and 3G5 (lower panel). F, localization of the 3G5 antibody epitope within the MDM2 hydrophobic pocket (pocket shown in ribbon representation, colored gold). Nutlin-3 bound to the pocket in stick representation (chlorine in green, carbon in gray, nitrogen in blue, and oxygen in red). The epitope of 3G5 antibody consists of four critical residues; Leu66, Tyr67, Asp68, and Glu69, surrounded by trypsin cleavable Lys and Arg residues (highlighted in purple). This figure was prepared using PyMOL (W. L. DeLano (2002) PyMOL, DeLano Scientific, San Carlos, CA), and structural data were from file 1TTV-RCSB PDB.