Abstract
We report a novel isoform of non-muscle myosin II-C (NM II-C), NM II-C2, that is generated by alternative splicing of an exon, C2, encoding 41 amino acids in mice (33 in humans). The 41 amino acids are inserted into loop 2 of the NM II-C heavy chain within the actin binding region. Unlike most vertebrate non-muscle and smooth muscle myosin IIs, baculovirus-expressed mouse heavy meromyosin (HMM) II-C2 demonstrates no requirement for regulatory myosin light chain (MLC20) phosphorylation for maximum actin-activated MgATPase activity or maximum in vitro motility as measured by the sliding actin filament assay. In contrast, noninserted HMM II-C0 and another alternatively spliced isoform HMM II-C1, which contains 8 amino acids inserted into loop 1, are dependent on MLC20 phosphorylation for both actin-activated MgATPase activity and in vitro motility (Kim, K. Y., Kovacs, M., Kawamoto, S., Sellers, J. R., and Adelstein, R. S. (2005) J. Biol. Chem. 280,22769 -22775). HMM II-C1C2, which contains both the C1 and C2 inserts, does not require MLC20 phosphorylation for full activity similar to HMM II-C2. These constitutively active C2-inserted isoforms of NM II-C are expressed only in neuronal tissue. This is in contrast to NM II-C1 and NM II-C0, both of which are ubiquitously expressed. Full-length NM II-C2-GFP expressed in COS-7 cells localizes to filaments in interphase cells and to the cytokinetic ring in dividing cells.
Mammalian non-muscle myosin IIs (NM IIs)2 belong to the conventional Class II myosins and are hexameric proteins composed of two heavy chains and two pairs of light chains, referred as the 20-kDa regulatory myosin light chain (MLC20) and the 17-kDa essential myosin light chain (MLC17). These myosins self-associate through their tail regions to form bipolar filaments that pull on actin filaments to produce force to drive important cellular functions such as cytokinesis, cell polarity, and cell migration (1-4). Three isoforms of the non-muscle myosin heavy chain (NMHC), II-A, II-B, and II-C, have been identified in vertebrates. They are products of three different genes, MYH9 (5, 6), MYH10 (6), and MYH14 (7, 8), respectively, in humans. It is well established that the enzymatic activity of these myosins is regulated by phosphorylation of MLC20, which is catalyzed by a number of enzymes, including myosin light chain kinase (MLCK), and Rho kinase (9-14).
Alternative splicing of pre-mRNA of NMHC II genes generates multiple mRNAs to enhance protein diversity in the NM II family. Work from this laboratory and others (8, 15-18) has established that both NMHC II-B and II-C undergo alternative splicing to generate several isoforms. In the case of NMHC II-B, 10 amino acids are incorporated into loop 1 at amino acid 212 (NMHC II-B1), and 21 amino acids are inserted into loop 2 at amino acid 622 (NMHC II-B2; see Ref. 15). These isoforms have been expressed as proteins, and their biochemical and functional importance has been studied extensively (19-22). Recently, it has been reported that baculovirus-expressed heavy meromyosin (HMM) II-B2 lacks actin-activated MgATPase activity and cannot propel actin filaments in an in vitro motility assay following MLC20 phosphorylation (22) even though HMM II-B0 and II-B1 show normal phosphorylation-dependent activities (21). These two inserted isoforms (NM II-B1 and NM II-B2) are only expressed in neuronal tissues, and the results of ablating each of them and NM II-B in mice have been reported (23-25).
For NMHC II-C, an alternative exon encoding 8 amino acids is incorporated into loop 1 at amino acid 227 (NMHC II-C1) at a location homologous to that of the B1 insert. Unlike NMHC II-B1, which is only expressed in neuronal tissue, NMHC II-C1 is found in a variety of tissues such as liver, kidney, testes, brain, and lung (8). The presence of the C1 insert in baculovirus-expressed HMM II-C1 increases both the actin-activated MgATPase activity and in vitro motility of HMM II-C1 compared with HMM II-C0, the noninserted form. The activity of both HMM II-C0 and HMM II-C1 is dependent on MLC20 phosphorylation (26). NM II-C1 has been shown to be expressed in a number of tumor cell lines, and decreasing its expression using small interfering RNA delays a late step in cytokinesis in the lung tumor cell line A549 (27).
In this study, we report that an exon encoding 41 amino acids can be incorporated into loop 2 near the actin binding region at amino acid 636 of NMHC II-C in mice. Expression of NM II-C2 is limited to neural tissue in mice. We used the baculovirus system to express all four isoforms of HMM II-C and found that inclusion of the 41 amino acids in loop 2 results in an HMM with an actin-activated MgATPase activity and in vitro motility that are independent of MLC20 phosphorylation.
EXPERIMENTAL PROCEDURES
Identification of the C2 Insert in Mouse and Human NMHC II-C—Total RNA from various mouse tissues was isolated using the RNeasy minikit (Qiagen, Valencia, CA). Total RNA from human tissues was obtained from Clontech. 1 μg of isolated total RNA was reverse-transcribed using random hexamers and the Gene-Amp RNA PCR core kit (Applied Biosystems, Branchburg, NJ). The resulting cDNA was amplified by PCR using the primer sets flanking the C2 inserted region as follows: for mouse forward primer, 5′-CAG CGC CCC AGG AAC CTG CG-3′, and reverse primer, 5′-GCT CCA GGG CCT GGA TCA TCT T-3′; and for human forward primer, 5′-GGA TCA GGC CGA CTT CAG TG-3′, and reverse primer, 5′-GTT CCA GCG CCT GGA TCA TCT-3′. The PCR program included four cycles of denaturation at 94 °C for 1 min, annealing at 65 °C for 1 min, extension at 72 °C for 1 min, and then 31 cycles of denaturation at 94 °C for 30 s, annealing at 60 °C for 30 s, and extension at 72 °C for 30 s. RNA samples subjected to cDNA synthesis in the absence of reverse transcriptase were used as a negative control for genomic DNA contamination. Products generated by RT-PCR were analyzed on a 1.8% agarose gel. The slower migrating bands (694 bp for mouse and 650 bp for human) from cerebellum were extracted from the gel and subcloned into the pCR2.1-TOPO vector for sequencing using the TOPO TA cloning kit (Invitrogen), which identified the inserted isoform of both mouse and human NMHC II-C.
Construction of Baculovirus Expression Vectors—Recombinant HMM-like proteins of mouse NMHC II-Cs were expressed in the baculovirus/Sf9 system. The cDNAs (nucleotides 1-4071 encoding 1357 amino acids for II-C0; nucleotides 1-4095 encoding 1365 amino acids for II-C1; nucleotides 1-4194 encoding 1398 amino acids for II-C2; and nucleotides 1-4218 encoding 1406 amino acids for II-C1C2) (GenBank™ accession numbers for mouse NMHC II-C0, II-C1, and II-C2 are AY205605, AY363100, and EF602040, respectively) were used to create HMM II-C fragments. Aforesaid mouse primers were used to amplify the C2 insert-containing fragment from mouse cerebellum cDNA. The PCR fragment was digested with HincII and BspEI enzymes and cloned into HMM II-C0 and HMM II-C1 constructs (26) at HincII/BspEI sites to generate HMM II-C2 and HMM II-C1C2, respectively. Nucleotides encoding a FLAG epitope (DYKDDDDK) followed by a stop signal were appended to aid in purification. The HMM constructs were subcloned into the baculovirus transfer vector pFastBac1 (Invitrogen).
GFP-tagged Full-length cDNA Constructs of NMHC II-C2 and NMHC II-C1C2—The C2 insert and both the C1 and C2 insert-containing fragments were retrieved from pFastBac1-HMM II-C2 and pFastBac1-HMM II-C1C2 respectively, and separately cloned into the previously reported pEGFP-NMHC II-C0 (27) construct at the SphI/BstXI site.
Preparation and Purification of HMM II-C Isoforms—Recombinant HMMs of all four alternatively spliced isoforms of NMHC II-C were expressed in the baculovirus/Sf9 system along with the appropriate light chains (21) and purified as reported previously (28). In brief, virus containing heavy chain and virus containing both light chains were cotransfected into 3 × 109 Sf9 cells. After 72 h of growth, infected Sf9 cells were harvested by sedimentation and stored at -80 °C. The partially thawed pellet was extracted with 0.5 m NaCl, 10 mm MOPS (pH 7.3), 10 mm MgCl2, 1 mm EGTA, 3 mm NaN3, 2 mm ATP, 0.1 mm phenylmethylsulfonyl fluoride, 0.1 mm dithiothreitol, 5 μg/ml leupeptin, and protease inhibitor mixture (Sigma) after homogenization in a ground glass homogenizer and purified by FLAG affinity chromatography. Proteins eluted from the FLAG column (Sigma) were concentrated using a Sepharose Q column (Amersham Biosciences). The protein concentration was determined by Bradford assay using smooth muscle HMM as a standard.
ATPase Assay—Actin-activated MgATPase activities were measured by an NADH-linked assay as described by Wang et al. (29) in a buffer containing 10 mm MOPS (pH 7.0), 2 mm MgCl2, 0.1 mm EGTA, 1 mm ATP, 0.2 mm CaCl2, 1 μm calmodulin, with and without 10 μg/ml MLCK, 40 units/ml lactate dehydrogenase, 200 units/ml pyruvate kinase, 1 mm phosphoenolpyruvate, and 0.2 mm NADH, various concentrations of actin (1-30 μm), and HMM II-C isoforms (0.2-0.8 μm) at 25 °C. Actin filaments were stabilized by a 1.5-fold molar excess of phalloidin (Calbiochem). Data were corrected for the background ATPase activity of actin. To determine the kinetic constants Vmax and KATPase, the data were fitted to the Michaelis-Menten mathematical equation using OriginLab 7.5 (Microcal Corp., Northampton, MA). Reported means and S.D. are those of three to four separate experiments.
In Vitro Motility Assay—Motility assays were performed at 30 °C in a buffer comprising 50 mm KCl, 20 mm MOPS (pH 7.4), 5 mm MgCl2, 0.1 mm EGTA, 1 mm ATP, 50 mm dithiothreitol, 0.7% methylcellulose, 2.5 mg/ml glucose, 0.1 mg/ml glucose oxidase, and 2 μg/ml catalase. All HMM II-C isoforms were introduced at a protein concentration of 0.2 mg/ml into a flow chamber with a nitrocellulose-coated coverslip. The surface was subsequently blocked by 1 mg/ml bovine serum albumin and then incubated for 1 min at room temperature in a solution containing 5 μm unlabeled F-actin, 1 mm ATP, 0.2 mm CaCl2, 1 μm calmodulin, with and without 4 μg/ml MLCK. After wash-out, 20 nm F-actin labeled with rhodamine-phalloidin (Molecular Probes, Carlsbad, CA) in the above assay buffer was applied to the flow chamber. The velocity of actin filament movement over the myosin-coated surface was analyzed using the Cell-Trak system (Motion Analysis, Santa Rosa, CA) as described earlier (28). Reported means and S.D. are those of three separate experiments.
Electrophoresis, Immunoblotting, and Immunoprecipitation—Extracts of Neuro 2A cells were prepared for SDS-PAGE as described previously (27). Briefly, cells on tissue culture plates were washed twice with cold phosphate-buffered saline and directly lysed with Laemmli sample buffer. Proteins were separated by SDS-PAGE on 4-20% polyacrylamide gradient Tris-glycine gels, transferred to a polyvinylidene difluoride membrane (Invitrogen), and blocked in 5% nonfat milk and 0.1% Tween 20 in phosphate-buffered saline. The blot was incubated with antibodies to the C2 sequence (0.27 μg/ml) at 4 °C overnight, washed, and then incubated with horseradish peroxidase-conjugated secondary antibodies (Pierce) at room temperature for 1 h, and treated with SuperSignal West Femto luminol enhancer solution (Pierce). Luminescence signal was captured on Biomax MR film (Eastman Kodak Co.). Antibodies to GFP (Sigma) were used to compare expression of GFP-tagged NMHC II-Cs. Tissue extracts from mouse brain were prepared using a buffer composed of 80 mm MOPS (pH 7.4), 150 mm NaCl, 10 mm MgCl2, 5 mm ATP, 4 mm EDTA, 1 mm dithiothreitol, 1% Nonidet P-40, and protease inhibitors (chymostatin, 1-chloro-3-tosylamido-7-amino-2-heptanone, l-(tosylamido-2-phenyl)-ethyl chloromethyl ketone, leupeptin, phenylmethylsulfonyl fluoride, aprotinin, benzamidine, and pepstatin A), at 4 °C. The lysates were sedimented at 10,000 × g for 10 min, and the supernatant was subjected to immunoprecipitation with antibody specific to the C terminus of NMHC II-C as reported previously (8). The immunoprecipitates were fractionated by SDS-PAGE on 6% polyacrylamide Tris-glycine gels, and NMHC II-C2 was detected with antibody specific to the C2 insert following the above method. Protein concentrations were determined using a Bio-Rad protein assay kit. Glycerol-urea polyacrylamide gels were run according to Perrie and Perry (30) and silver-stained as per the manufacturer (Sigma).
Preparation and Characterization of Antibodies—A peptide of 12 amino acids (EQGGLQQFTLLG) located at the very N terminus of the mouse C2 insert was synthesized and used for antibody generation in rabbits. After the 3rd boost, serum was passed through aforesaid peptide column to affinity purify the antibody.
Cell Culture, Transfection, and Immunofluorescence Microscopy—Mouse Neuro 2A cells were obtained from ATCC (Manassas, VA) and grown in Eagle's minimum essential medium containing 10% fetal bovine serum. For transient transfection, 1 μg of plasmid DNA/ml of culture medium was transfected using Effectene® transfection reagents (Qiagen). COS-7 cells (ATCC) were grown on chamber slides in high glucose Dulbecco's modified Eagle's medium containing 10% fetal bovine serum. The NMHC II-C2-GFP construct or II-C0-GFP construct was transiently transfected into COS-7 cells using Nucleofector R kits (Amaxa, Cologne, Germany) following the manufacturer's instructions. Forty eight hours after DNA transfection, cells were washed, fixed, and permeabilized as described previously (31). The samples were blocked with phosphate-buffered saline containing 0.1% bovine serum albumin and 5% normal goat serum for 30 min at room temperature followed by incubation with rhodamine-labeled phalloidin at room temperature for 2 h. Nuclei were counterstained with 4′, 6-diamidino-2-phenylindole from Invitrogen. The images were collected using a Zeiss LSM 510 meta confocal microscope.
RESULTS
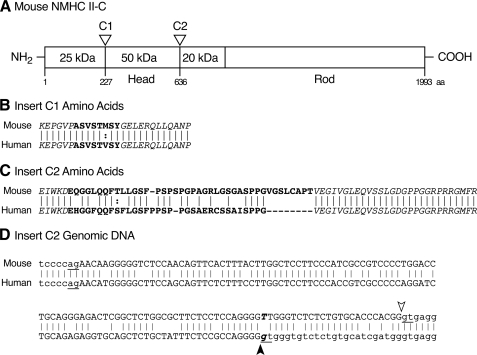
Loop 2 of NMHC II-C Has an Insert—Golomb et al. (8) reported that an exon encoding an 8-amino acid insert, C1, can be inserted into loop 1, which is near the ATP binding region of NMHC II-C. Here, we report that another exon encoding 41 amino acids in mice and 33 amino acids in humans, C2, can be incorporated into loop 2 within the actin binding region (Fig. 1). Insertion of C1 and C2 inserts occurs because of the alternative splicing of pre-mRNA transcribed from the Myh14 gene. Fig. 1 illustrates the position and sequence of both the C1 and C2 inserts in the heavy chain of NM II-C. Note that the overlapping amino acid residues of C2 are 67% identical between humans and mice and that there is complete identity of amino acids in the flanking regions between humans and mice (Fig. 1C). A comparison of the mouse and human genomic sequences flanking the C2 exon provides information as to why the mouse insert is larger than the human (Fig. 1D). Although the human genomic sequence contains a donor splice site (gt) following the 4 guanine bases as indicated by the filled arrowhead, the mouse sequence does not (TT instead of gt, Fig. 1D). The mouse sequence does contain a donor splice site 24 nucleotides 3′ to the human one (Fig. 1D, gt, open arrowhead). Although the human gene also has a potential donor splice site at the same position as the mouse gene, so far we have no evidence for splicing at this site in human mRNA.
FIGURE 1.
Diagram of the location of inserted sequences in NMHC II-C. A, schematic diagram of mouse NMHC II-C shows the location of the C1 insert (near the ATP binding region) and C2 insert (near the actin binding region). The 25-50- and the 50-20-kDa boundaries designate proteolytic sensitive regions where loop 1 and loop 2 are located. B, amino acid sequences of the C1 insert (boldface letters) shown for mice and humans are from Golomb et al. (8). The flanking sequences of the C1 insert, shown in italics, are conserved between mice and humans. C, mouse NMHC II-C2 has 41 amino acids (boldface letters) inserted in loop 2, whereas human C2 has 33 amino acids (boldface letters). Similar to C1, the sequences (italics) flanking the C2 insert are conserved between mice and humans (GenBank™ accession numbers for mouse NMHC II-C2 and human NMHC II-C2 are EF602040 and FJ041910, respectively). Dashes are introduced to maximize alignment between amino acids. D, genomic sequence of mouse and human NMHC II-C (Myh 14) shows the C2 exon and flanking introns. GenBank™ accession numbers for mouse and human genomic DNA sequences are NT_039424 and NT_011109, respectively. Capital letters represent the exon sequence, and lowercase letters are intron sequence. Arrowheads and underlined ag and gt nucleotides indicate the splice sites. The mismatch of boldface italic letters (T and g) results in different usage of the splice donor sites between mice (open arrowhead) and humans (filled arrowhead). Note that exon C2 starts with the second nucleotide of the codon for glutamic acid, GAA. Lines connect identical amino acids or nucleotides between mouse and human sequences.:, conservative amino acid substitution; aa, amino acid.
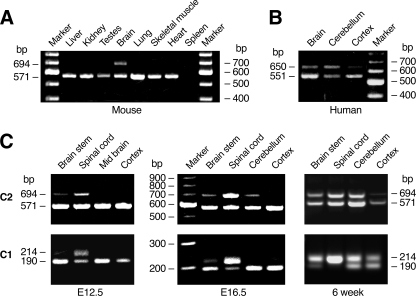
Neuron-specific Expression of NMHC II-C2—RT-PCR analysis using specific primers flanking the C2 insert was performed to determine the expression pattern of NMHC II-C2 in a number of tissues. Although the PCR product lacking the C2 insert (571 bp) is detected in most tissues except spleen, expression of the C2 insert (694 bp) in mice was confined to the brain (Fig. 2A). This is in contrast to NMHC II-C1, which is expressed in a variety of tissues and cell lines (8, 27). Expression of C2 inserted mRNA is also detected in human brains as shown by the generation of a 650-bp fragment compared with 551 bp for the mRNA lacking C2 (Fig. 2B). Fig. 2C illustrates the expression of the C1 and C2 inserted mRNAs in various parts of the brain at embryonic day (E) E12.5 and E16.5 and at 6 weeks of age in mice. Expression of the C2 and C1 insert increases from E12.5 to adulthood; however, the time course of the increase and the extent of increase in the different parts of the brain and spinal cord are somewhat different (Fig. 2C).
FIGURE 2.
Expression of C2-inserted mRNAs in mouse and human brain tissues. A, RT-PCR was used to analyze the indicated tissue from 6-week-old mice for C2 insert expression. Primers were chosen to flank the inserted C2 sequence (see “Experimental Procedures”). B, RT-PCR product for the C2 insert regions from adult human brain tissues. C, expression of C2 insert (upper panels) and C1 insert (lower panels) in indicated parts of mouse brain at E12.5, E16.5, and 6 weeks of age.
The results shown for the spinal cord at E16.5 and 6 weeks and the brain stem at 6 weeks for the C1 and C2 inserts imply that some of the mRNA must contain both the C1 and C2 inserts. To detect mRNAs that contain both the C1 and C2 inserts, we carried out RT-PCR using primers located in both inserts. We succeeded in obtaining the expected 1,353-bp product for the spinal cord, brain stem, and cerebellum, confirming the presence of mRNAs with both inserts at 6 weeks (data not shown). Thus all four possible mRNA variants encoding NMHC II-C0, -C1, -C2, and -C1C2 appear to be present in the mouse brain.
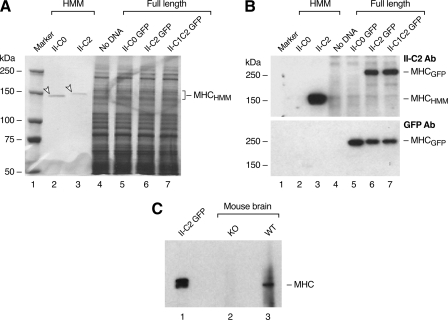
NMHC II-C2 and NMHC II-C1C2 Proteins Are Detected by an Antibody—In an effort to detect NMHC II-C2/C1C2 protein expression, we used the first 12 residues of the mouse C2 insert sequence (EQGGLQQFTLLG) to generate antibodies in rabbits that were then affinity-purified using the same peptide. Fig. 3A shows a Coomassie Blue-stained gel with the following samples: baculovirus expressed HMM encompassing residues 1-1357 of noninserted HMM II-C0 (26), baculovirus expressed HMM II-C2 (see below), and extracts of the Neuro 2A cell line transfected as labeled with plasmids encoding full-length NMHC II-C fused to GFP. Fig. 3B is an immunoblot showing that the antibody detects the myosin heavy chain of HMM II-C2 (lane 3) but not HMM II-C0 (lane 2). It also detects NMHC II-C2-GFP and NMHC II-C1C2-GFP but not NMHC II-C0-GFP (Fig. 3B, lanes 5-7). The lower panel in Fig. 3B uses an antibody to GFP to compare II-C-GFP expression. To detect endogenous NMHC II-C2/C1C2 in brain, we used an antibody raised to the C-terminal sequence of NMHC II-C to immunoprecipitate II-C isoforms from brain extracts prepared from wild-type and NM II-C ablated mice.3 Fig. 3C is an immunoblot probed with the antibody to the C2 insert and shows that the C2 insert antibody recognizes a band migrating close to 230 kDa in the wild-type brain extract (lane 3) but not in the brain extract from II-C ablated mice (lane 2). Neuro 2A cells transfected with the NMHC II-C2-GFP plasmid served as a positive control (lane 1). Therefore, the mouse brain expresses the C2-inserted NMHC-II-C as a protein.
FIGURE 3.
NMHC II-C2 and NMHC II-C1C2 are detected by specific antibodies. A, Coomassie Blue-stained gel of samples used in immunoblot shown in B. Each lane is designated above the gel lane: lane 1, protein markers; lanes 2 and 3, baculovirus-expressed HMM II-C isoforms (arrowheads); lanes 4-7, extracts of Neuro 2A cells either untransfected or transfected with plasmids encoding full-length NMHC II-C isoforms fused to GFP at the C terminus. B, immunoblots from a gel similar to that shown in A. The upper panel was probed with the C2-specific antibody (Ab), and the blot was stripped and reprobed with antibody to GFP (lower panel). C, immunoblot probed with the C2-specific antibody. Lane 1, lysate of Neuro 2A cells transfected with NMHC II-C2-GFP (positive control). Lanes 2 and 3 are immunoprecipitates from NMHC II-C knock-out (KO) and wild-type (WT) mouse brains, respectively, using an antibody specific to the C-terminal sequence of NMHC II-C for immunoprecipitation. MHC, myosin heavy chain.
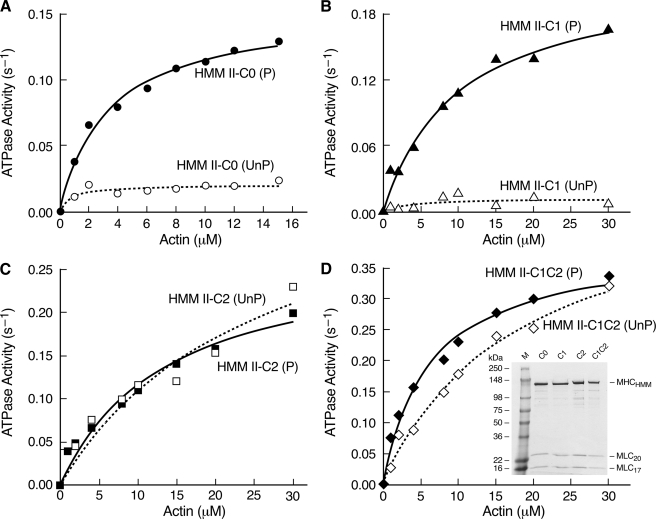
Expression of Recombinant FLAG-tagged HMM II-C Isoforms—To characterize and compare the biochemical and motile properties of the various NM II-C isoforms, NM II-C0, II-C1, II-C2, and II-C1C2, we used the baculovirus expression system to generate HMM II-C for the various spliced isoforms. Fig. 4D (inset) is a Coomassie Blue-stained SDS-polyacrylamide gel showing each of the purified HMM II-C heavy chains together with both light chains. Of note, both HMM II-C2 and II-C1C2 heavy chains migrate slightly more slowly than HMM II-C0 and II-C1 heavy chains because of the additional 41 amino acids. Although the results for the biochemical and motile activity of HMM II-C0 and II-C1 have been published (26), we thought it was important to express and characterize all four isoforms simultaneously because of our unusual findings for HMM II-C2 and HMM II-C1C2 (see below).
FIGURE 4.
Expression of FLAG-tagged HMM II-C isoforms and actin-activated MgATPase activities. Actin-activated MgATPase activities of HMM II-C0 (A), HMM II-C1 (B), HMM II-C2 (C), and HMM II-C1C2 (D) were measured at 25 °C with various concentrations of actin before (open symbol, dotted line) and after (closed symbol, solid line) phosphorylation by MLCK. Data sets were fitted to a hyperbolic equation to determine the kinetic constants, Vmax and KATPase (see Table 1). The data shown are from a single representative preparation of each HMM. UnP, unphosphorylated; P, phosphorylated. Of note is that in contrast to HMM II-C0 or C1, both HMM II-C2 and C1C2 have similar MgATPase activities in the phosphorylated and unphosphorylated states. D, inset, Coomassie Blue-stained gel of purified HMM II-C proteins. C0, HMM with no insert; C1, HMM with C1 insert; C2, HMM with C2 insert; C1C2, HMM with C1 and C2 inserts. The spurious, slower migration of MLC20 in the SDS gel is because of the buffer system and the particular markers used in this gel (Invitrogen SeeBlue® Plus2 Pre-stained Standards).
Actin-activated MgATPase Activity of HMM II-C Isoforms—To date, the actin-activated MgATPase activity of practically all vertebrate NM IIs and smooth muscle myosin II (SM II) is regulated by phosphorylation of MLC20. These myosins have a very low actin-activated MgATPase activity when their light chain is unphosphorylated, and this activity increases upon phosphorylation of MLC20 on Ser-19/Thr-18. We measured the basal MgATPase activity (in the absence of actin) of each of the HMM II-C proteins as well as the actin-activated MgAT-Pase activity before and after MLC20 phosphorylation using an NADH-coupled assay at 25 °C at low ionic strength over a range of actin concentrations (Fig. 4, A-D). The data for each HMM was fitted to a hyperbolic equation to determine the kinetic constants Vmax and KATPase (actin concentration at half-maximal activation, see Table 1). The data demonstrate a major difference among the activities of the isoforms in the absence of MLC20 phosphorylation. The C2-inserted isoforms, both C2 and C1C2, show more than 9-10-fold higher values for Vmax compared with the isoforms lacking C2 (C0 and C1, Table 1). Following MLC20 phosphorylation with MLCK, the Vmax values of the C0 and C1 isoforms increase ∼5-fold compared with the unphosphorylated state, whereas those of the isoforms containing C2 and C1C2 remain similar or decrease somewhat. All four isoforms have Vmax values in a similar range. These results demonstrate that the C2-inserted HMMs (C2 and C1C2) do not require MLC20 phosphorylation for full activation (Fig. 4, C and D, and Table 1).
TABLE 1.
Actin-activated MgATPase activity of HMM II-C before and after phosphorylation by MLCK
Actin-activated MgATPase activity was measured at 25 °C as described under “Experimental Procedures.” The values for Vmax and KATPase are the mean and S.D. from three to four experiments from three different protein preparations. II-C0, noninserted HMM II-C; II-C1, inserted HMM II-C with an 8-amino acid insert in loop 1; II-C2, inserted HMM II-C with a 41-amino acid insert in loop 2; II-C1C2, inserted HMM II-C with both C1 and C2 inserts; basal MgATPase is measured without actin.
|
HMMs |
Basal MgATPase, Vmax |
Actin-activated MgATPase |
|||
|---|---|---|---|---|---|
|
−MLCK |
+MLCK |
||||
| Vmax | KATPase | Vmax | KATPase | ||
| s−1 | s−1 | μm | s−1 | μm | |
| II-C0 | <0.006 | 0.03 ± 0.01 | 1.2 ± 0.7 | 0.16 ± 0.02 | 4.6 ± 1.6 |
| II-C1 | <0.007 | 0.04 ± 0.03 | 15.6 ± 13.3 | 0.21 ± 0.01 | 10.9 ± 4.6 |
| II-C2 | <0.04 | 0.34 ± 0.09 | 13.9 ± 6.9 | 0.27 ± 0.06 | 7.2 ± 5.5 |
| II-C1C2 | <0.02 | 0.36 ± 0.09 | 12.9 ± 5.4 | 0.26 ± 0.06 | 3.8 ± 1.7 |
In Vitro Motility—An important functional property of myosin is its ability to translocate actin filaments. This is quantified using the in vitro motility assay with rhodamine-phalloidin-labeled actin. The movement of actin filaments over the myosin-coated surface was monitored by fluorescence microscopy, and the sliding speed was calculated using the CellTrak system. As Table 2 shows, in the absence of MLC20 phosphorylation, there was no significant movement (<0.01 μm/s) of actin filaments bound to HMM II-C0- and II-C1-coated surfaces. Following MLC20 phosphorylation, as expected, the actin filaments were propelled by HMM II-C0 and II-C1 with an average velocity of 0.04 and 0.09 μm/s, respectively, similar to a previous report (26). In contrast, HMM II-C2 and II-C1C2 can translocate actin filaments with a velocity of 0.06 μm/s whether or not MLC20 is phosphorylated. These results indicate that the insertion of 41 amino acids into loop 2 renders HMM II-C2s constitutively active with respect to in vitro motility.
TABLE 2.
In vitro motility of HMM II-C with and without phosphorylation by MLCK
In vitro motility was measured at 30 °C as described under “Experimental Procedures.” The values are the mean and S.D. of three experiments from two different protein preparations. II-C0, noninserted HMM II-C; II-C1, inserted HMM II-C with an 8-amino acid insert in loop 1; II-C2, inserted HMM II-C with a 41-amino acid insert in loop 2; II-C1C2, inserted HMM II-C with both C1 insert in loop 1 and C2 insert in loop 2; NSM, no significant movement.
|
HMMs |
In vitro motility |
|
|---|---|---|
| −MLCK | +MLCK | |
| μm/s | ||
| II-C0 | NSM | 0.04 ± 0.005 |
| II-C1 | NSM | 0.09 ± 0.004 |
| II-C2 | 0.06 ± 0.001 | 0.06 ± 0.003 |
| II-C1C2 | 0.06 ± 0.004 | 0.06 ± 0.003 |
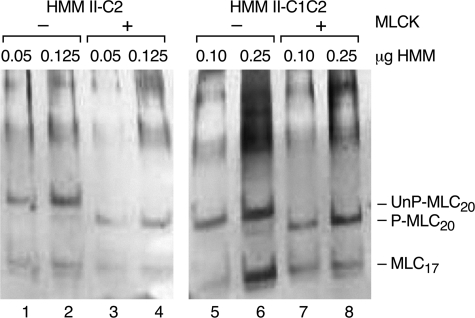
Phosphorylation Status of Purified HMM II-C—Although unlikely, the possibility existed that HMM II-C2 and HMM II-C1C2 might be phosphorylated on MLC20 during or prior to isolation and purification from baculovirus-infected Sf9 cells. To address this possibility we analyzed both HMMs using a glycerol-urea polyacrylamide gel, which separates the phosphorylated and unphosphorylated MLC20 on the basis of charge. Fig. 5 shows that as purified, MLC20 is not significantly phosphorylated. The very lightly stained band migrating with P-MLC20 of HMM II-C1C2 in Fig. 5, lane 6, shows that less than 5% of the MLC20 is phosphorylated. Following treatment with MLCK, essentially all MLC20 becomes phosphorylated as shown by the more rapid migration of MLC20 for both HMM II-C2 and II-C1C2 in the glycerol gel (P-MLC20). This confirmed that MLC20 was not phosphorylated as isolated. Thus we conclude that the C2-inserted HMMs are constitutively active with respect to both actin-activated MgATPase activity and in vitro motility.
FIGURE 5.
Phosphorylation of MLC-20. Silver-stained glycerol-urea polyacrylamide gels of purified HMM II-C2 and HMM II-C1C2 are shown. In this system, the unphosphorylated (UnP) MLC20 migrates more slowly than phosphorylated (P) MLC20. The amount of HMM protein loaded is shown above each lane. HMM II-C2 and II-C1C2 were incubated in the presence (lanes 3, 4 and 7, 8) or absence (lanes 1, 2 and 5, 6) of MLCK.
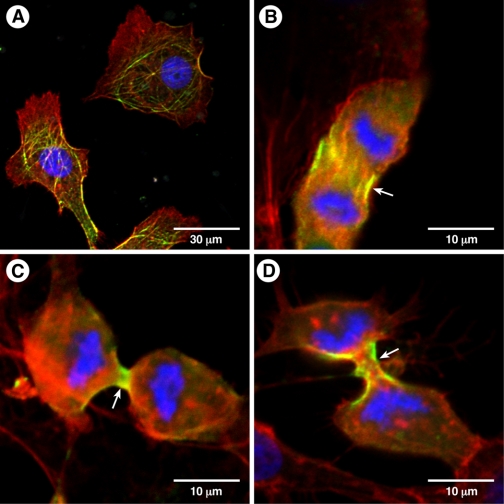
Localization of NM II-C2-GFP in COS-7 Cells—COS-7 cells, a kidney cell line cloned from monkeys, are known to contain mostly NM II-B (>95%) and small amounts of NM II-C (31). Because we were unable to identify a cell line that contained NM II-C2, we transfected a plasmid encoding NM II-C2-GFP or a second plasmid encoding NM II-C0-GFP into COS-7 cells to localize the distribution of the isoforms in interphase cells and during cytokinesis. Fig. 6A shows that NM II-C2-GFP forms filamentous structures, some of which colocalize with actin filaments in interphase cells. Similar to other NM IIs, NM II-C0-GFP (Fig. 6B) and NM II-C2-GFP (Fig. 6, C and D) localized to the contractile ring during cytokinesis. Thus the presence of a rather large insert in the heavy chain of NMHC II-C does not appear to interfere with its cellular function and localization.
FIGURE 6.
Localization of NM II-C2-GFP in COS-7 cells. COS-7 cells were transfected with a plasmid encoding full-length NMHC II-C2-GFP or NMHC II-C0-GFP. Interphase (A) or dividing cells (B-D) are shown. Actin filaments were detected with rhodamine-labeled phalloidin (red). 4′, 6-Diamidino-2-phenylindole was used for nuclear staining. Note that NM II-C2-GFP localizes to filaments in interphase cells (A) and to the contractile ring in a dividing cell (C and D, arrow). NM II-C0-GFP also localizes to the contractile ring (B, arrow).
DISCUSSION
In this study we characterize a novel, alternatively spliced isoform of NM II-C with an unusually large 41-amino acid insert in the actin binding region of the myosin heavy chain. This insert is only expressed in neural tissue. The alternative exon encoding this insert is expressed in both humans and mice, but the human exon encodes a peptide insert that terminates eight amino acids prior to that found in the mouse because of a difference in splice donor sites (Fig. 1D). Unlike most vertebrate NM II and SM II isoforms characterized to date, HMM II-C2 and II-C1C2 do not require phosphorylation of their regulatory light chain at Ser-19/Thr-18 for activity, as measured by actin-activated MgATPase activity and by the sliding actin in vitro motility assay.
In contrast to these constitutively active isoforms of myosin II, Kim et al. (22) describe an alternatively spliced isoform of NM II-B, NM II-B2, whose MgATPase activity is not activated by actin and does not propel actin filaments even when Ser-19 is phosphorylated. The C2 and B2 alternative exons are spliced into homologous locations in loop 2 of the NMHC, and similar to NMHC II-C2, NMHC II-B2 is also found to be expressed only in neural tissue (15, 16, 25). Because of its low activity we speculated that NM II-B2 plays a role in cellular functions that depend more on the structural properties, rather than the motor activity of myosin II. Interestingly, when the NM II-B2 isoform was specifically ablated in mice, Ma et al. (25) found that the cerebellar Purkinje cells had decreased numbers of spines and branches and that some of these cells were abnormally mislocalized in the cerebellum. These mice also walked with a staggered gait reflecting cerebellar defects. We have not yet studied the in vivo function of NM II-C2, but Alhopuro et al. (32) identified unregulated mutant forms of SM II in patients suffering from an intestinal neoplasia. The mutations were found in the first segment of the tail region of the myosin heavy chain (amino acid 1044) and in the globular head (amino acid 501), and resulted in a constitutively active SM II with a 2-fold reduction in Vmax and in vitro motility. Two other alternatively spliced isoforms, NM II-C1 and NM II-B1, which result from inserted alternative exons in loop 1 of NMHC II-C and NMHC II-B, respectively, are regulated by MLC20 phosphorylation, and the presence of the insert acts to increase the actin-activated MgATPase activity and in vitro motility of the NM IIs (21, 26). These isoforms differ in their distribution profile in mouse and human tissues in that expression of NM II-B1, similar to NM II-C2, is limited to neuronal tissue, whereas that of NM II-C1 is ubiquitous in its distribution among mouse and human tissues (8, 15, 27). Including both the C1 and the C2 inserts in HMM results in a myosin that, similar to HMM II-C2, is constitutively active. Based on the unusual regulatory properties of NM II-B2 and II-C2, we may conclude that isoforms that result from alternative splicing at loop 2 are more diverse in biochemical and functional properties than those with inserts in loop 1.
Although we were able to demonstrate, using RT-PCR, that mRNA encoding NMHC II-C2 (and NMHC II-C1C2) is expressed in different regions of the mouse brain such as the brain stem, cerebellum, and spinal cord, our efforts to detect the inserted isoform using a C2-specific antibody have been less successful to date. We have not been able to detect the presence of this isoform in a number of neuronal cell lines such as PC 12, Neuro 2A, or Y-79 or in brain tissues using immunofluorescence microscopy (data not shown). On the other hand we were able to detect the isoform by immunoblot using the C2-specific antibody following immunoprecipitation of NMHC II-C from whole brain lysates. This suggests that the isoform may only be present in limiting quantities or that we have not located a specific area of the brain that contains it. Alternatively, this may reflect the apparent lack of antigenicity of the peptide used to generate antibodies. Further work, such as specific ablation of the isoform in mice, could shed more light on its in vivo function. Of note is the ability of full-length NM II-C2-GFP to localize to filaments following transfection into COS-7 cells and to the cytokinetic ring in dividing COS-7 cells. Similar to phosphorylation-dependent NM IIs, NM II-C2-GFP was capable of rescuing multinucleation in COS-7 cells treated with small interfering RNA to NM II-B (Ref. 31 and data not shown). This observation is similar to that of Dean and Spudich (33) who used a constitutively active Drosophila NM-II substituted with a phosphomimetic MLC20 and showed that it could be recruited to the contractile ring and support cytokinesis.
A number of investigators, including Wendt et al. (34), Burgess et al. (35), and Jung et al. (36), have offered a possible explanation for the structural basis of the inactive forms of unphosphorylated SM II, NM II, and invertebrate striated muscle. They have shown that the inactive form adopts a compact “switched off” conformation in solution as detected by either cryo- or negatively stained electron microscopy coupled with single-particle image analysis. In this conformation the loop 2 region of one head (termed the blocked head) is in contact with the converter region of the other (free) head in an asymmetric arrangement. The tail folds at two points (one fold is near amino acid 1198 and other is at amino acid 1591, in the case of SM II). Together these conformational changes result in a much more compact molecule. The blocked head is limited in its movement because of extensive interactions with the tail, whereas the free head is more mobile. Phosphorylation on MLC20 by MLCK breaks the intramolecular interaction between heads and leads to an open, unfolded conformation that is able to manifest biochemical properties such as actin-activated MgATPase activity and actin sliding activity (35, 36).
One possible explanation for our results from the kinetic and actin sliding assays is that both HMM II-C2 and HMM II-C1C2 remain in the “active form” (i.e. open and unfolded) even in the unphosphorylated state. The C2 insert, which is 41 amino acids in length, may disturb the interactions between the two heads or between the blocked head and the first segment of the tail, keeping both heads free for interaction with actin and for hydrolyzing ATP, which is necessary for contractile activity.
Studies from different laboratories have demonstrated that changing the net charge of loop 2, the length of loop 2, or replacing loop 2 with a loop 2 from an unregulated myosin II, can alter the regulation of SM II. For example, Rovner et al. (37) replaced loop 2 in SM II with that of skeletal and cardiac myosin II. They found that the chimeric myosins no longer required phosphorylation of MLC20 for activity. Another study showed that removal of the N-terminal 12 amino acids of loop 2 or addition of lysine residues at the C terminus of loop 2 caused an increase in activity in phosphorylated and unphosphorylated SM II, while decreasing the degree of regulation. In contrast, replacing the C-terminal 16 amino acids of SM II loop 2 (leaving the N-terminal 12 amino acids) with skeletal muscle loop 2 caused a decrease in activity and gain in the degree of regulation by phosphorylation (38). These studies suggested that both the N-terminal 12 amino acids and charge intensity at the C terminus of loop 2 regulate SM II activity. On the other hand insertion of the B2 insert into loop 2 of NM II-C0, which abrogates the activity of NM II-B0, did not significantly alter myosin activity nor did it interfere with its dependence on MLC20 phosphorylation for activity (22). In this study, incorporation of the C2 insert, which is 41 amino acids in length and has no net charge, led to a constitutively active form of NM II-C. Further studies will be necessary to learn whether this unusually large insert imposes a new form of regulation on NM II-C or simply does not require an off state for its physiological activity. Identifying other kinases or signaling pathways that can switch off this molecule would enhance our understanding of the role of this myosin in neuronal tissue.
Acknowledgments
We thank the members of the Laboratory of Molecular Cardiology and Laboratory of Molecular Physiology for reagents and helpful discussions. We thank Christian A. Combs and Daniela A. Malide (Light Microscopy Core Facility, NHLBI, National Institutes of Health) for professional skills and advice. We thank Antoine Smith and Dalton Saunders for skillful technical assistance. We also thank Xuefei Ma for help with Fig. 6 and Mary Anne Conti for kindly and carefully reading the manuscript.
The nucleotide sequence(s) reported in this paper has been submitted to the Gen-Bank™/EBI Data Bank with accession number(s) EF602040 and FJ041910.
This work was authored, in whole or in part, by National Institutes of Health staff.
Footnotes
The abbreviations used are: NM II, non-muscle myosin II which includes both heavy and light chains; SM II, smooth muscle myosin II; E, embryonic day; NMHC, non-muscle myosin heavy chain; HMM, heavy meromyosin; MLC20, 20-kDa myosin light chain; MLC17, 17-kDa myosin light chain; MLCK, myosin light chain kinase; MOPS, 4-morpholinepropanesulfonic acid; RT, reverse transcription; GFP, green fluorescent protein.
X. Ma, S. Kawamoto, and R. S. Adelstein, unpublished data.
References
- 1.Conti, M. A., Kawamoto, S., and Adelstein, R. S. (2008) in Myosins: A Superfamily of Molecular Motors (Coluccio, L. M., ed) Vol. 7, pp.223 -264, Springer, Dordrecht, The Netherlands [Google Scholar]
- 2.Berg, J. S., Powell, B. C., and Cheney, R. E. (2001) Mol. Biol. Cell 12780 -794 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bresnick, A. R. (1999) Curr. Opin. Cell Biol. 1126 -33 [DOI] [PubMed] [Google Scholar]
- 4.Conti, M. A., and Adelstein, R. S. (2008) J. Cell Sci. 12111 -18 [DOI] [PubMed] [Google Scholar]
- 5.Toothaker, L. E., Gonzalez, D. A., Tung, N., Lemons, R. S., Le Beau, M. M., Arnaout, M. A., Clayton, L. K., and Tenen, D. G. (1991) Blood 781826 -1833 [PubMed] [Google Scholar]
- 6.Simons, M., Wang, M., McBride, O., Kawamoto, S., Yamakawa, K., Gdula, D., Adelstein, R. S., and Weir, L. (1991) Circ. Res. 69530 -539 [DOI] [PubMed] [Google Scholar]
- 7.Leal, A., Endele, S., Stengel, C., Huehne, K., Loetterle, J., Barrantes, R., Winterpacht, A., and Rautensttauss, B. (2003) Gene (Amst.) 312165 -171 [DOI] [PubMed] [Google Scholar]
- 8.Golomb, E., Ma, X., Jana, S. S., Preston, Y. A., Kawamoto, S., Shoham, N. G., Goldin, E., Conti, M. A., Seller, J. R., and Adelstein, R. S. (2004) J. Biol. Chem. 2792800 -2808 [DOI] [PubMed] [Google Scholar]
- 9.Kamm, K. E., and Stull, J. T. (2001) J. Biol. Chem. 2764527 -4530 [DOI] [PubMed] [Google Scholar]
- 10.Matsumura, F. (2005) Trends Cell Biol. 15371 -377 [DOI] [PubMed] [Google Scholar]
- 11.Poperechnaya, A., Varlamova, O., Lin, P. J., Stull, J. T., and Bresnick, A. R. (2000) J. Cell Biol. 151697 -708 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Amano, M., Ito, M., Kimura, K., Fukata, Y., Chihara, K., Nakano, T., Matsuura, Y., and Kaibuchi, K. (1996) J. Biol. Chem. 27120246 -20249 [DOI] [PubMed] [Google Scholar]
- 13.Burridge, K., and Wennerberg, K. (2004) Cell 116167 -179 [DOI] [PubMed] [Google Scholar]
- 14.Sandquist, J. C., Swenson, K. I., Demali, K. A., Burridge, K., and Means, A. R. (2006) J. Biol. Chem. 28135873 -35883 [DOI] [PubMed] [Google Scholar]
- 15.Itoh, K., and Adelstein, R. S. (1995) J. Biol. Chem. 27014533 -14540 [DOI] [PubMed] [Google Scholar]
- 16.Takahashi, M., Hirano, T., Uchida, K., and Yamagishi, A. (1999) Biochem. Biophys. Res. Commun. 259 29-33 [DOI] [PubMed] [Google Scholar]
- 17.Kawamoto, S. (1996) J. Biol. Chem. 27117613 -17616 [PubMed] [Google Scholar]
- 18.Nakahata, S., and Kawamoto, S. (2005) Nucleic Acids Res. 332078 -2089 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Rosenfeld, S. S., Xing, J., Chen, L., and Sweeney, H. L. (2003) J. Biol. Chem. 27827449 -27455 [DOI] [PubMed] [Google Scholar]
- 20.Takahashi, M., Takahashi, K., Hiratsuka, Y., Uchida, K., Yamagishi, A., Uyeda, T. Q., and Yazawa, M. (2001) J. Biol. Chem. 2761034 -1040 [DOI] [PubMed] [Google Scholar]
- 21.Pato, M. D., Sellers, J. R., Preston, Y. A., Harvey, E. V., and Adelstein, R. S. (1996) J. Biol. Chem. 2712689 -2695 [DOI] [PubMed] [Google Scholar]
- 22.Kim, K., Kawamoto, S., Bao, J., Sellers, J. R., and Adelstein, R. S. (2008) Biochem. Biophys. Res. Commun. 369124 -134 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Tullio, A. N., Accili, D., Ferrans, V. J., Yu, Z.-X., Takeda, K., Grinberg, A., Westphal, H., Preston, Y. A., and Adelstein, R. S. (1997) Proc. Natl. Acad. Sci. U. S. A. 9412407 -12412 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Tullio, A. N., Bridgman, P. C., Tresser, N. J., Chan, C. C., Conti, M. A., Adelstein, R. S., and Hara, Y. (2001) J. Comp. Neurol. 43362 -74 [DOI] [PubMed] [Google Scholar]
- 25.Ma, X., Kawamoto, S., Uribe, J., and Adelstein, R. S. (2006) Mol. Biol. Cell 172138 -2149 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kim, K., Kovacs, M., Kawamoto, S., Sellers, J. R., and Adelstein, R. S. (2005) J. Biol. Chem. 28022769 -22775 [DOI] [PubMed] [Google Scholar]
- 27.Jana, S. S., Kawamoto, S., and Adelstein, R. S. (2006) J. Biol. Chem. 28124662 -24670 [DOI] [PubMed] [Google Scholar]
- 28.Wang, F., Harvey, E. V., Conti, M. A., Wei, D., and Sellers, J. R. (2000) Biochemistry 395555 -5560 [DOI] [PubMed] [Google Scholar]
- 29.Wang, F., Kovacs, M., Hu, A., Limouze, J., Harvey, E. V., and Sellers, J. R. (2003) J. Biol. Chem. 27827439 -27448 [DOI] [PubMed] [Google Scholar]
- 30.Perrie, W. T., and Perry, S. V. (1970) Biochem. J. 11931 -38 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Bao, J., Jana, S. S., and Adelstein, R. S. (2005) J. Biol. Chem. 28019594 -19599 [DOI] [PubMed] [Google Scholar]
- 32.Alhopuro, P., Phichith, D., Tuupanen, S., Sammalkorpi, H., Nybondas, M., Saharinen, J., Robinson, J. P., Yang, Z., Chen, L. Q., Orntoft, T., Merklin, J. P., Jarvinen, H., Eng, C., Moeslein, G., Houlaton, R. S., Lucassen, A., Tomlinson, I. P., Launonen, V., Ristimaki, A., Arango, D., Karhu, A., Sweeney, H. L., and Aaltosen, L. A. (2008) Proc. Natl. Acad. Sci. U. S. A. 1055513 -5518 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Dean, S. O., and Spudich, J. A. (2006) PLoS ONE 1e131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wendt, T., Taylor, D., Trybus, K. M., and Taylor, K. (2001) Proc. Natl. Acad. Sci. U. S. A. 984361 -4366 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Burgess, S. A., Yu, S., Walker, M. L., Hawkins, R. J., Chalovich, J. M., and Knight, P. J. (2007) J. Mol. Biol. 3721165 -1178 [DOI] [PubMed] [Google Scholar]
- 36.Jung, H. S., Komatsu, S., Ikebe, M., and Craig, R. (2008) Mol. Biol. Cell 193234 -3342 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Rovner, A. S., Freyzon, Y., and Trybus, K. M. (1995) J. Biol. Chem. 27030260 -30263 [DOI] [PubMed] [Google Scholar]
- 38.Rovner, A. S. (1998) J. Biol. Chem. 27327939 -27944 [DOI] [PubMed] [Google Scholar]