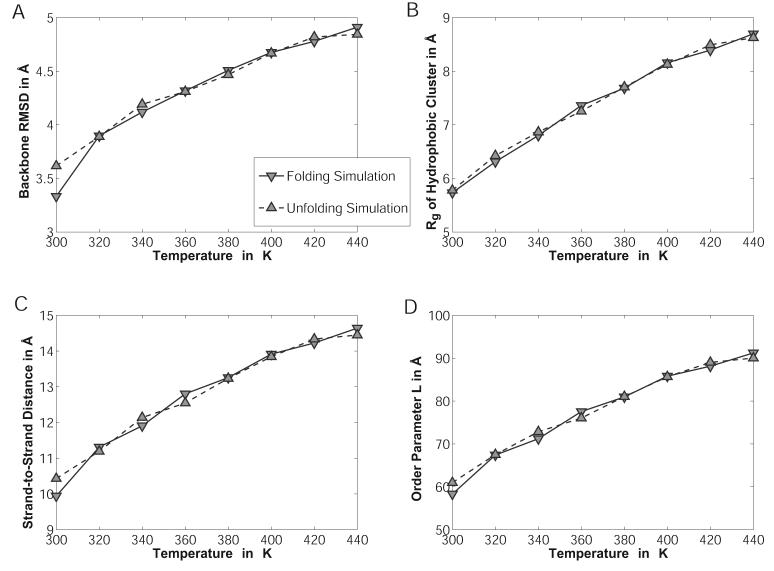
Figure 10.
The temperature dependence of various order parameters characterizing the simulated ensembles of the tryptophan zipper “trpzip1”. Sets of folding and unfolding simulations are shown as solid and dashed lines, respectively. Panel A shows the heavy backbone atom RMSD to the PDB structure (Model 1 in 1LE0) excluding the N-terminal serine and the C-terminal amide cap. Panel B shows the radius of gyration of the sidechains of the four tryptophan residues. Panel C shows the mean strand-to-strand distance for the perfect hairpin, whereas the order parameter L as defined by Snow et al.74 is shown in Panel D.