Figure 7.
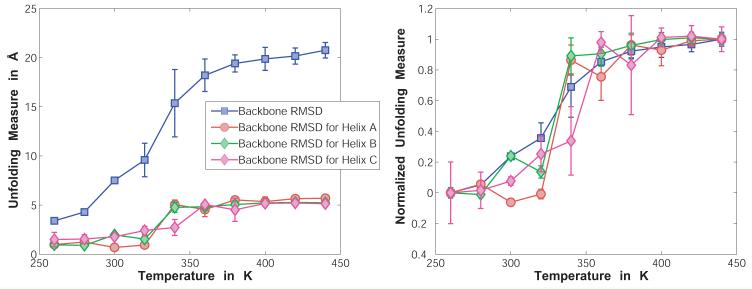
Unfolding measures for the engrailed homeodomain as a function of simulation temperature. Panel A shows four raw values for the RMSD to the PDB structure, which are based on all heavy backbone atoms excluding terminal residues as well as based on the heavy backbone atoms for the three helices individually. Using the PDB-numbering (1ENH) the helices were defined as residues 11 to 25 (A), residues 29 to 41 (B), as well as residues 43 to 57 (C). Likewise to Figure 6, the RMSD is based on structural alignments using only the corresponding residues as alignment criteria. Panel B shows values for the four measures normalized to their end points at 260K (0.0, fully folded) as well as 440K (1.0, fully unfolded). Error bars are obtained through block averaging, using a block size of 5×105 MC steps.