Abstract
Cathepsins have been best characterized in tumorigenesis and cell death and implicated in liver fibrosis; however, whether cathepsins directly regulate hepatic stellate cells (HSC) activation and proliferation, hence modulating their fibrogenic potential is largely unknown. Here we show that expression of cathepsin B (CtsB) and cathepsin D (CtsD) is negligible in quiescent HSC but parallels the increase of α-SMA and TGF-β during in vitro mouse HSC activation. Both cathepsins are necessary for HSC transdifferentiation into myofibroblasts, as their silencing or inhibition decreased HSC proliferation and the expression of phenotypic markers of HSC activation, with similar results observed with the human HSC cell line LX2. CtsB inhibition blunted AKT phosphorylation in activated HSC in response to PDGF. Moreover, during in vivo liver fibrogenesis by CCl4 administration, CtsB expression increased in HSC but not in hepatocytes, and its inactivation mitigated CCl4-induced inflammation, HSC activation and collagen deposition. Altogether, these findings support a critical role for cathepsins in HSC activation, suggesting that the antagonism of cathepsins in HSC may be of relevance for the treatment of liver fibrosis.
Keywords: HSC transdifferentiation, AKT phosphorylation, liver fibrosis
Liver fibrosis represents the final common pathological outcome of many chronic liver insults, and a major medical problem (1, 2). Despite recent advances in the understanding of the activation of hepatic stellate cells (HSC) and their role in liver fibrogenesis, relatively little is known about the molecular mechanisms that regulate these processes.
As in other tissues, the fibrotic component of the liver’s wound-healing response is mediated by myofibroblasts. In the injured liver, myofibroblasts are potentially derived from a number of cellular sources, including activated HSC, which constitute a predominant reservoire for myofibroblasts (1-4). HSC are responsible for extensive synthesis and deposition of extracellular matrix (ECM) during liver fibrosis. Upon liver injury, HSC undergo a phenotypic transformation from non-proliferating, retinoid storing cells, to a proliferating, matrix producing phenotype (1-6). HSC activation occurs in a two-step process, the initiation phase characterized by loss of retinoids and upregulation of cytoskeletal protein expression such as α -SMA and desmin, and the perpetuation phase where the ensuing myofibroblasts not only produce almost all of the ECM but also a broad array of cytokines and chemokines that amplify HSC activation via autocrine stimulation (1, 6). Up to date, treatment for advanced liver fibrosis and cirrhosis is inefficient, and the development of successful anti-fibrotic therapies is imperative.
The cathepsins family comprises the catalytic serine (cathepsin G), aspartate (cathepsins D and E) and cysteine (cathepsins B, C, H, F, K, L, O, S, V, and W) peptidases that exhibit endo-or exopeptidase activities (7, 8). Apart from their known role in protein turnover, recent data have revealed that cathepsins can also regulate multiple processes including neovascularization of endothelial progenitor cells (9), antigen presentation (10), cell growth and tissue homeostasis (11-13). Moreover, cathepsins also have important functions outside lysosomes. For example, execution of programmed cell death, degradation of ECM components, or induction of invasive cell growth have been described when cathepsins are released into the cytosol or secreted in the extracellular space (14-18). Consistent with these functions, and since ECM degradation is a hallmark of tumorigenesis, aspartic protease cathepsin D (CtsD) and the cysteine cathepsin B (CtsB) have been implicated in cancer progression, invasion and metastasis (7, 14, 19-21).
In addition, cathepsins have also been involved in liver injury and fibrogenesis. For instance, cathepsin T and CtsD activation has been observed in experimental fibrogenesis (22) and previous studies reported high levels of CtsB, D and L in serum of patients with cirrhosis and hepatocellular carcinoma (23-25). Moreover, using two-dimensional polyacrylamide gel electrophoresis, Kristensen et al. reported the upregulation of a number of proteins, including CtsD, during the activation of rat HSC and liver fibrogenesis (26), whose expression correlated with PDGF receptor β levels during HSC activation (27). Finally, Canbay et al described a lysosomal pathway of hepatocellular apoptosis and liver injury following bile duct ligation (BDL) in mice, in which CtsB played a key role, suggesting that CtsB-mediated hepatocellular apoptosis contributed to inflammation and fibrogenesis (28). However, despite these investigations whether cathepsins (CtsB or D) directly regulate HSC activation and proliferation, or if modulation of cathepsins expression in HSC impacts on their fibrogenic potential has not been specifically addressed.
Thus, our aim was to examine the contribution of CtsB and CtsD in the activation of mouse HSC in vitro and in the development of fibrosis in vivo following CCl4 treatment. We show that the levels of CtsB and CtsD are negligible in quiescent HSC but increase in parallel with the upregulation α-SMA and TGF-β during HSC transdifferentiation into myofibroblasts. Genetic silencing or pharmacological inhibition of cathepsins mitigate HSC activation and hence progression of liver fibrogenesis.
EXPERIMENTAL PROCEDURES
Isolation and culture of hepatic stellate cells
C57BL/6 mice 8-12 week old were from Charles River. All animals received humane care according to the criteria outlined in the “Guide for the Care and Use of Laboratory Animals”. Hepatic stellate cells (HSC) were isolated from C57BL/6 mice by perfusion with collagenase-pronase as described (29) with small modifications. HSC were separated from parenchymal cells by 60×g centrifugation, collecting the supernatant for centrifugation at 450×g for 10min. Pellet or non-parenchymal cells were resuspended and purified over a 17.2% Hystodenz density gradient by centrifugation. The cloudy strip was collected and the HSC were cleaned with Krebs-Henseleit buffer by centrifugation of 450×g for 10 minutes. Cells were cultured in DMEM complemented with 10% FBS, and antibiotics at 37°C in a humidified atmosphere of 95% air and 5% CO2. Culture purity was assessed by retinoid autofluorescence. Mouse HSC were not passaged and were used from day-2 to day-10.
In vivo liver fibrogenesis
C57BL/6 mice were treated with carbon tetrachloride (CCl4) at a dose of 5μL (10% CCl4 in corn oil)/g body weight, by intraperitoneal injection for 6 weeks twice a week. One hour before treatment with CCl4, and during the last four weeks, mice received either CtsB inhibitor (Ca074Me), or vehicle. Stock solutions of Ca074Me were made at a concentration of 10mg/ml in dimethyl-sulfoxide. The stock was diluted 1:10 in saline and administered at 10mg/kg body weight by intraperitoneal injection. Control animals received vehicle alone.
CtsB and CtsD activities
CtsB activity was assayed fluorimetrically with Z-Arg-Arg-7-amido-4-methylcoumarin hydrochloride (60μmol/L) at pH 7.4 and 37 °C as previously described (30). Briefly, the assay buffer employed contained 20mmol/L HEPES pH 7.4, 5% Sucrose, 0.1% 3-[(3-Cholamidopropyl)dimethylammonio]-1-propanesulfonate, 2mmol/L EDTA, 5mmol/L dithiothreitol, and 2mmol/L cysteine. The fluorimetric assay (λex: 360 nm; λem: 460 nm) was performed in 96-well plate using 20 μg of protein per sample. Similarly, CtsD activity was determined fluorimetrically (λex: 400 nm; λem: 505 nm) using the specific substrate N-Acetyl-Arg-Gly-Phe-Phe-Pro-7-amido-4-trifluoromethylcoumarin (60μmol/L) at pH 7.4 and 37 °C. The assay buffer contained 20mM HEPES pH 7.4, 5% Sucrose, 0.1% 3-[(3-Cholamidopropyl)dimethylammonio]-1-propanesulfonate, 2mM EDTA, and 5mM dithiothreitol. Results were expressed as cathepsin activity (slope of fluorescence emission after 40 min) per mg of protein.
In vitro siRNA transfection
To silence CtsB and CtsD expression, specific pre-designed siRNAs for mouse were used for transfection using Lipofectamine LTX and PLUS following the manufacters’ instruction. Briefly, 100nmol/L siRNA, 5μL of PLUS and 200μL of Optimem were mixed for 15min at room temperature. 6μL of Lipofectamine LTX were added afterwards, transferring the mixture to a 6-well plate after 25 min. In some cases, cells were transfected with both siRNA against CtsB and CtsD, evaluating the expression of α-SMA, TGF-β and 2’-5’ oligoadenylate synthetase 1 (OAS1). Cells were assayed usually 48h after siRNAs transfection.
[3H] Thymidine incorporation
Proliferation was estimated as the amount of [3H] thymidine incorporated into TCA-precipitable material. HSC were cultured in a 6-well plate, and transfected at day 5. [3H] thymidine (1μCi/mL) was added at 12, 24 and 48h after transfection and incubated for 24h. The reaction was stopped by addition of 2ml cold TCA (5%). After two rinses with cold TCA (5%), the radioactivity incorporated into TCA-insoluble material was recovered with 2.5mL of 0.5 N NaOH and counted by liquid scintillation counting.
Statistical analyses
All images display representative data from at least 3 independent observations. Statistical analyses were performed using Microsoft Excel software. The experiments were repeated at least three times. The statistical significance of differences was performed using the unpaired, non-parametric Student’s t test.
Reagent, Antibodies, and Methods
RESULTS
CtsB and CtsD expression during mouse HSC activation
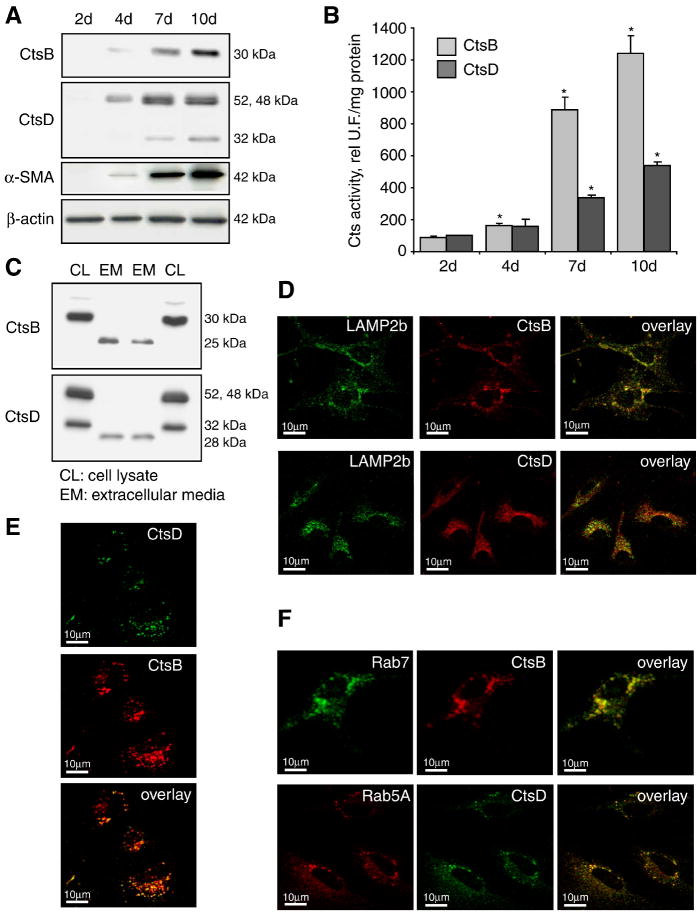
Since the culture of HSC is known to induce its transdifferentiation into a myofibroblast-like phenotype, freshly mouse primary HSC were isolated to analyze CtsB and CtsD expression at different time-points during culture. As shown in Fig. 1A the expression of CtsB and CtsD was almost negligible in quiescent HSC (day 2); however, their levels markedly increased during the time of culture with a maximum expression of both cathepsins seen at days 7-10; moreover, its increase over time paralleled the appearance of α-SMA (Fig. 1A), an indicator of the phenotypic transformation of HSC into myofibroblasts. The higher protein expression observed in panel A was accompanied by increased functional activity of both cathepsins (Fig. 1B). More important, conditioned media from overnight cultures of 7 day-old HSC revealed the presence of CtsB and CtsD in the extracellular media (Fig. 1C). This is consistent with the study by Kristensen et al., which reported the presence of CtsD in the extracellular media from activated rat HSC cultures (26). As illustrated in Fig. 1C, there is a molecular weight shift in the extracellular media for both cathepsins compared to the form present in cellular lysates. In the case of the secreted form of CtsB, the molecular weight is 25 kDa, derived from the processing of the 30kDa intracellular active form (31). Unexpectedly, the molecular weight of the secreted form of CtsD is approximately 28kDa, smaller than the intracellular 32kDa double-chain of mature CtsD form, which has not been described previously to the best of our knowledge. Moreover, both, CtsB and CtsD were localized in lysosomes, as shown by confocal imaging of 7-day old HSC double-stained with a lysosomal marker, and anti-CtsB or anti-CtsD antibodies (Fig. 1D). In addition, both cathepsins colocalized with each other in a punctate pattern (Fig. 1E), and co-localized also with Rab5A and Rab7, early and late endosome markers respectively, indicating that these enzymes follow the secretory pathway for its secretion to the extracellular media (Fig. 1F).
Figure 1.
Mouse HSC express CtsB and CtsD. (A) Time-course of CtsB, CtsD, and α-SMA protein expression by western blot. (B) CtsB and CtsD activity. (C) CtsB and CtsD present in the extracellular media (EM) differ from the intracellular form (CL). Western blot displaying two representative experiments (Experiment #1: lane 1-2; experiment #2: lane 3-4). (D-F) Confocal imaging of 7 day-old HSC displaying CtsB, CtsD, with a lysosomal marker (LAMP2b) (D), among themselves (E), and with early (Rab5A) or late (Rab11) endosome markers (F). Data are mean±SD, n=4 and *p≤ 0.05 vs. day 2 HSC. Scale bar 10μm (D-F).
CtsB and CtsD silencing downregulates mouse HSC proliferation and expression of fibrogenic genes
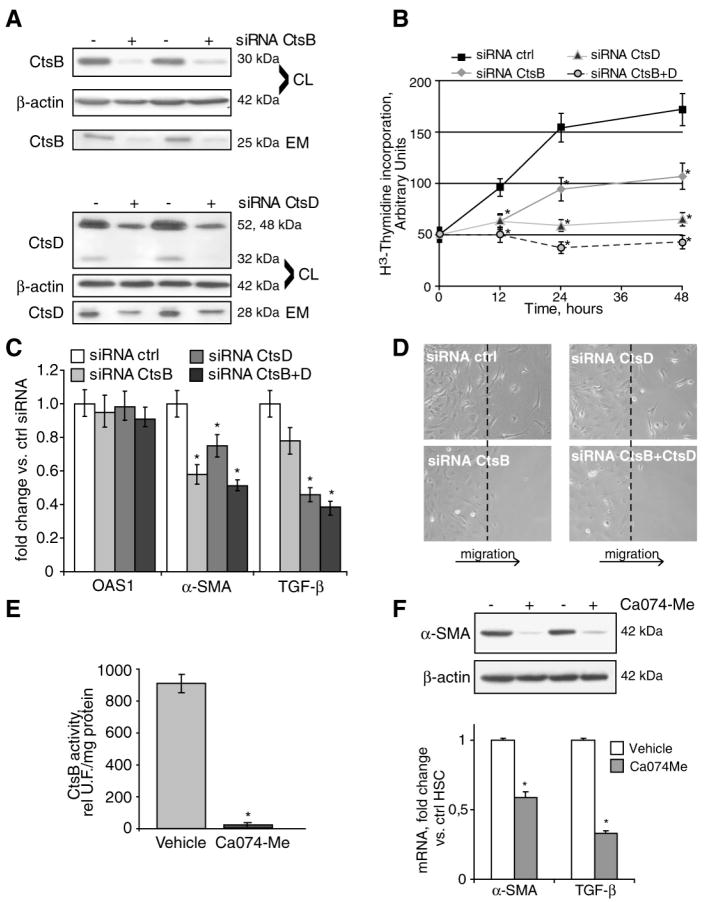
We next examined the contribution of CtsB and CtsD on HSC activation following CtsB/CtsD silencing via siRNAs. To this aim, 5-day old HSC were transfected with control, CtsB and/or CtsD siRNA. The targeting of CtsB and CtsD by siRNAs reduced their protein levels by more than 80% in both cell lysates and extracellular media (Fig. 2A). Silencing of either CtsB or CtsD resulted in decreased cell proliferation (Fig. 2B), compared to HSC transfected with control siRNA, with a more prominent effect observed for CtsD than CtsB. Blocking simultaneously both cathepsins resulted in a synergic potentiation of the inhibitory effect on proliferation (Fig. 2B). Moreover, downregulation of CtsB and/or CtsD also affected the levels of phenotypic markers of HSC activation, such as α-SMA and TGF-β, as soon as 24h after transfection, without affecting the interferon responsive gene 2’-5’-oligoadenylate synthetase 1 (OAS1) (32), indicating that siRNA transfection did not induce interferon response (Fig. 2C). As a positive control of IFN-response we used LPS (1μg/ml, 24h) resulting in 6-7 fold increase in OAS1 mRNA (not shown). In addition, we observed a dramatic loss of HSC migration, examined by a wound-healing assay, in cells depleted of CtsB, but not of CtsD, indicating a possible role of CtsB in the regulation of migration (Fig. 2D). We also examined the effect of pharmacological inhibition of CtsB on HSC activation. Parallel to the effect observed after silencing CtsB, its inhibition by Ca074Me, a potent and specific CtsB inhibitor (Fig. 2E) (33), resulted in the downregulation of α-SMA and TGF-β mRNA both at level of mRNA and protein (Fig. 2F).
Figure 2.
CtsB or CtsD silencing weakens HSC activation. CtsB and CtsD expression (A), proliferation (B), OAS1, α-SMA and TGB-β mRNA expression (C), and migration (D) after CtsB and/or CtsD silencing for 48h in 5 day-old HSC. CtsB activity (E) and α-SMA protein, and α-SMA and TGB-β mRNA expression (F) after Ca074Me challenge (10μmol/L) for 48h in 5-day old HSC. Data are mean±SD, in B and C, n=3 and *p≤ 0.05 vs. siRNA Ctrl transfected HSC; in E and F, n=3 and *p<0.001 vs. vehicle treated HSC.
Role of CtsB and CtsD in the human HSC cell line LX2
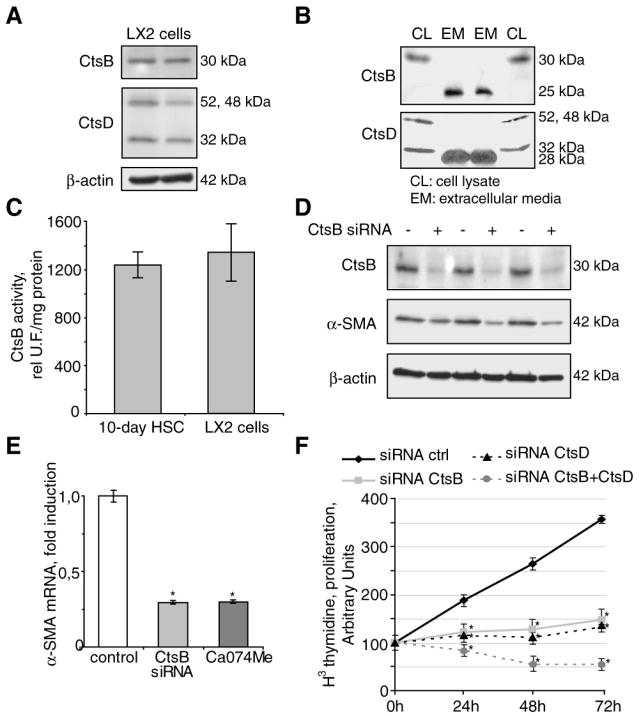
To examine the relevance of the preceding findings in human fibrogenesis, we next examined the expression of CtsB and CtsD in LX2 cells, an immortalized human HSC cell line similar to human activated HSC (34). As seen, LX2 exhibited the presence of both CtsB and CtsD, not only in the cellular lysates, but also in the extracellular media (Fig. 3A,B). In addition, CtsB activity in LX2 cells was similar to that of 10-day old mouse HSC cultures (Fig. 3C). Silencing of CtsB by siRNA transfection or its inhibition using Ca074Me downregulated α-SMA mRNA and protein levels in LX2 cells (Fig. 3D,E), while this effect was not observed upon CtsD silencing (not shown). However, reduction of CtsB and/or CtsD levels by siRNA decreased cell proliferation, compared to control siRNA-transfected LX2 cells (Fig. 3F). In contrast, CtsB inhibition by Ca074Me did not affect the expression of TGF-β mRNA in LX2 cells, nor their migration in a wound-healing assay (not shown), probably reflecting the fact that TGF-β regulation and migration in fully activated LX2 cells may be different compared to primary HSC, which models the initial phase of the transdifferentiation process (35, 36).
Figure 3.
CtsB and CtsD in LX2 cells. CtsB and CtsD expression by Western blot in LX2 cells (A), and its presence in the extracellular media (B). In B, the Western blot displays two representative experiments (Experiment #1: lane 1-2; experiment #2: lane 3-4). CtsB activity in LX2 cells and 10-day HSC (C). α-SMA protein (D), and mRNA expression (E) after CtsB silencing for 48h. (F) Proliferation after CtsB and/or CtsD silencing. Data are mean±SD, in D, n=3 and *p≤ 0.001 vs. Ctrl HSC; in E, n=3 and *p≤0.05 vs. siRNA Ctrl transfected LX2 cells.
Processing of CtsB and CtsD in mouse HSC
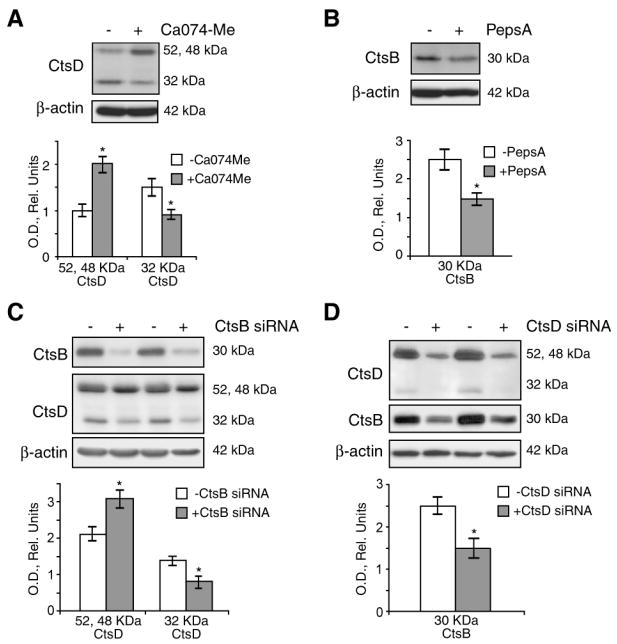
Cathepsins are synthesized as inactive precursors, which are then processed either autocatalytically or by other enzymes to remove an N-terminal pro-peptide (37). For instance, in mouse fibroblasts CtsD processing has been shown to require the function of cysteine cathepsins, such as CtsB, to generate a mature CtsD form (38). Conversely, CtsD has been shown to process CtsB precursor by the cleavage of the pro-cathepsin polypeptide (39). Thus, we examined the effect of CtsB/CtsD inhibition or silencing on their reciprocal processing in primary mouse HSC. As seen, CtsB inhibition by Ca074Me increased the CtsD precursor pro-form (52 and 48 kDa), while decreasing the mature form (32 kDa) (Fig. 4A). Similarly, after CtsD inhibition by Pepstatin A, an aspartyl protease inhibitor that blocks CtsD activity among other proteases (40), we observed the downregulation of the CtsB mature form (30 kDa) (Fig. 4B). Comparable results were obtained after silencing CtsB (Fig. 4C) and CtsD by siRNAs (Fig. 4D). Altogether, these results imply that CtsB and CtsD contribute to their mutual proteolytic processing in primary mouse HSC.
Figure 4.
CtsB and CtsD processing in HSC. Inhibition of CtsB in 5 day-old HSC with Ca074Me (10μmol/L) for 24h affects CtsD processing (A). Similarly, CtsD inhibition, with Pepstatin A (10μg/ml) for 24h, resulted in less CtsB active form (B). Parallel results were observed after CtsB or CtsD silencing (C,D). Data are mean±SD, *p≤ 0.05 vs. untreated HSC.
CtsB modulates the PI3K/AKT pathway in activated mouse HSC
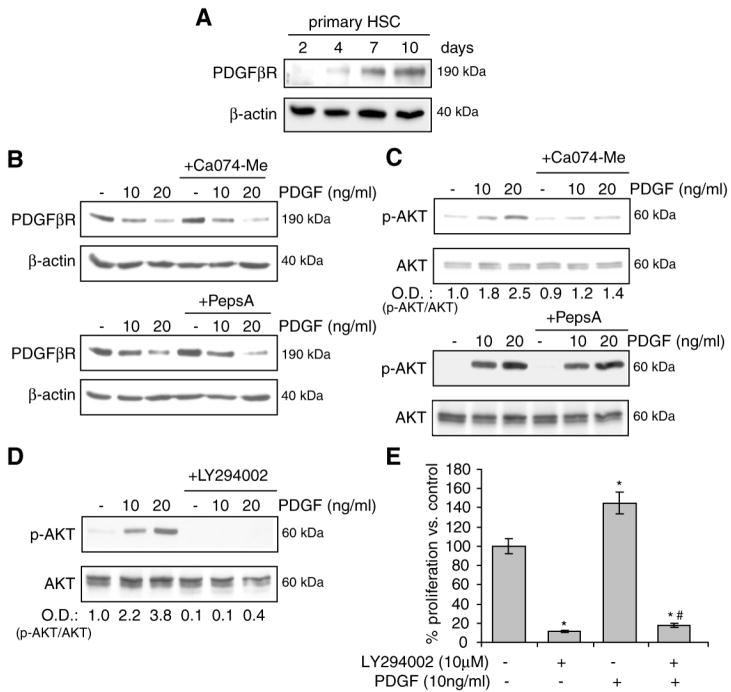
Platelet-derived growth factor-β receptor (PDGFβR) plays a key role in the transformation of HSC into myofibroblasts and has been suggested as a putative target of cathepsins in activated HSC (27, 41). Thus, to investigate if CtsB and CtsD modulate the expression of PDGFβR, we first examined their levels as a function of the activation of HSC from day-2 to day-10 in culture, and the effect of CtsB or CtsD inhibition on PDGFβR degradation after platelet-derived growth factor (PDGF) challenge. As observed (Fig. 5A), PDGFβR increased its expression during the activation of mouse HSC, in parallel to the expression observed for CtsB and CtsD (Fig. 1A). Pharmacological inhibition of CtsB or CtsD did not affect the degradation of PDGFβR induced by its ligand PDGF (Fig. 5B). However, while PDGF elicited a rapid phosphorylation of AKT, observed 15 min after PDGF challenge in activated HSC, this process was markedly reduced in HSC cultures supplemented with the CtsB inhibitor Ca074Me (Fig. 5C). Moreover, CtsD inhibition by Pepstatin A did not affect the phosphorylation of AKT induced by PDGF (Fig. 5C). Similar effects were observed after inhibition of the PI3K/AKT pathway by LY294002, which blocked AKT phosphorylation by PDGF (Fig. 5D) and decreased HSC proliferation following PDGF challenge (Fig. 5E). Overall, these findings suggest that CtsB regulates the mitogenic potential of HSC via modulation of AKT in response to PDGF.
Figure 5.
PI3K/AKT activation after CtsB inhibition in vitro. (A) PDGFβR levels during mouse HSC activation from day-2 to day-10. (B) Effect of CtsB (Ca074Me 10μmol/L, 24h) or CtsD (Pepstatin A 10μg/ml, 24h) inhibition on PDGFβR degradation after challenge with PDGF (60 min), and on AKT phosphorylation after PDGF challenge (15 min) (C). (D,E) Effect of the PI3-kinase inhibitor LY294002 (10μmol/L, 60 min preincubation) on the phosphorylation of AKT by PDGF (15 min) (D), and on the proliferation for 24h (E). Data are mean±SD, in D, n=3 and *p≤ 0.05 vs. control HSC; # p≤ 0.01 vs. PDGF-treated HSC.
CtsB overexpression in CCl4-treated mice
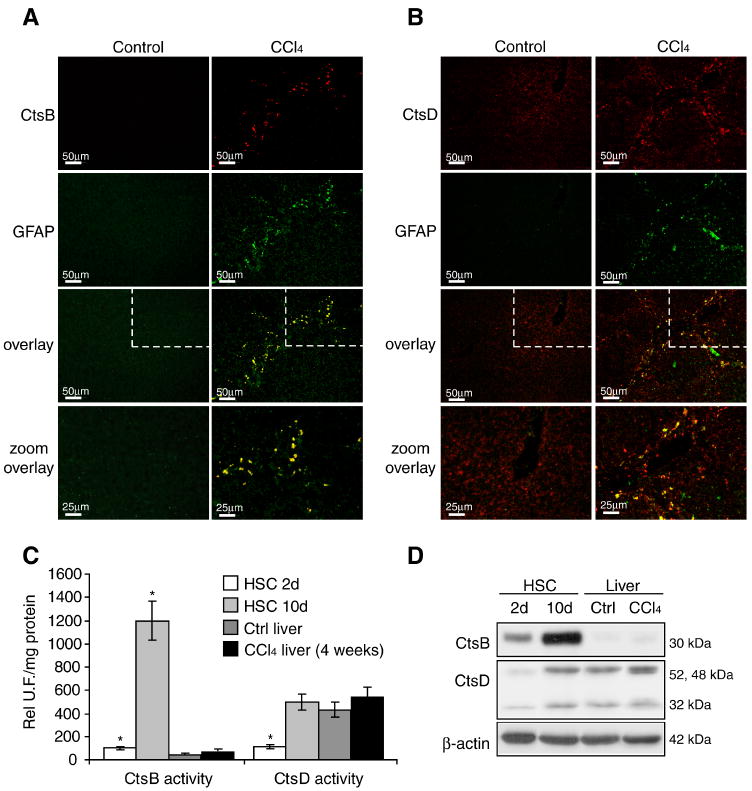
Since the preceding findings suggested a critical role of CtsB and CtsD in the activation of mouse primary HSC, we next examined their expression during in vivo liver fibrogenesis induced by CCl4, a well established model of liver injury. Immunohistochemical staining of liver sections displayed an intense staining of CtsB only in CCl4-treated mice that co-localized with glial fibrillary acidic protein (GFAP), an HSC marker (42) (Fig. 6A). In contrast, CtsD staining of liver sections exhibited a more diffuse pattern similar in control and CCl4 treated livers that partially co-localized with GFAP (Fig. 6B). Since these findings suggested that the increase expression of CtsB during liver fibrogenesis is restricted to HSC and because this particular cathepsin has been involved in TNF, cholestasis-mediated hepatocellular injury and free fatty acid-induced hepatic lipotoxicity in HepG2 cells (28, 43, 44), we examined the activity of CtsB and CtsD in CCl4-treated liver in comparison with that of 2-day and 10-day old mouse HSC cultures. While CtsD activity was similar in the liver of control or CCl4-treated mice, and in 10-day old HSC cultures, the activity of CtsB was very low in mouse liver, regardless of CCl4 treatment, compared to the activity observed in 10-day old HSC (Fig. 6C). Similar results were observed when analyzing CtsB and CtsD protein levels (Fig. 6D), confirming the immunohistochemical findings (Fig. 6A,B). Thus, these results indicate that CtsB and CtsD are differentially expressed in HSC and hepatocytes during CCl4-induced liver fibrogenesis, and that CtsB upregulation is limited to HSC in CCl4-treated mice.
Figure 6.
CtsB and CtsD expression in experimental fibrosis in vivo. (A,B) Immunofluorescence imaging of CtsB (A) or CtsD (B) and GFAP in control and 4-week treated CCl4 liver sections. The lower panel of A and B displays a magnification of the upper-right section indicated by the dashed line (C,D) CtsB, and CtsD activity (C) or protein expression (D) in day-2, day-10 HSC, and control and 4-weeks CCl4 treated livers. Data are mean±SD, in C, n=3 and *p≤ 0.01 vs. control liver. Scale bars 50μm in all panels, except in the lower panels (25μm).
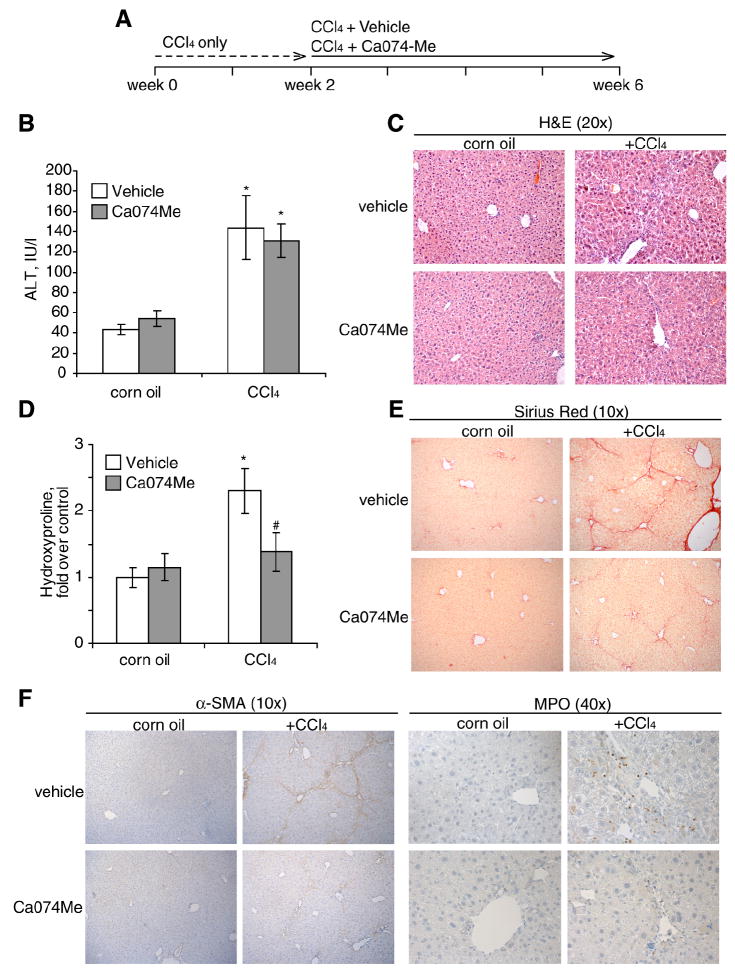
CtsB inhibition mitigates CCl4-mediated liver fibrogenesis
Given the above findings, we next asked whether CtsB inhibition could be of relevance in reversing liver fibrogenesis in vivo following CCl4 administration. In order to stimulate HSC activation before inhibiting CtsB, CCl4 was administered twice a week for 2 weeks, followed by CCl4 and Ca074Me administration for 4 additional weeks (Fig. 7A). CtsB inhibition by itself did not alter any of the parameters studied in control animals (Fig. 7). CCl4 challenge for 6 weeks increased serum ALT compared to control or vehicle levels, which was not significantly altered after CtsB inhibition by Ca074Me (Fig. 7B). H&E staining of liver sections (Fig. 7C) displayed ECM accumulation and sinusoidal dilatation in CCl4-treated mice, which were decreased after treatment with Ca074Me. These therapeutic effects were not due to impaired metabolism of CCl4 in Ca074Me-treated animals since microsomal cytochrome P450 2E1 activity was comparable in both CCl4 treated groups irrespectively of the presence of the inhibitor (not shown). However, CtsB inhibition in CCl4-treated mice resulted in a more preserved architecture of liver parenchyma with less fibrosis. Moreover, as shown in Fig. 7D, determination of the hydroxyproline content in the liver revealed a significant decrease in collagen accumulation in CCl4-treated mice that received Ca074Me, compared to mice challenged with CCl4 alone. This result was further confirmed by the increased detection of collagen fibers by Sirius Red staining (Fig. 7E) in CCl4-treated animals compared to those treated with CtsB inhibitor. In addition, CtsB inhibition in CCl4 treated animals decreased α-SMA positive staining of liver sections, accompanied by almost negligible neutrophilic infiltration as determined by myeloperoxidase staining (Fig. 7F). Thus, these findings indicate that while the function of CtsB in CCl4-induced hepatocellular damage is negligible, CtsB plays a critical role in in vivo liver fibrogenesis.
Figure 7.
CtsB inhibition in CCl4-treated mice. (A) Experimental setup of CCl4 and Ca074Me administration to mice. (B-F) 6-week after initial CCl4 challenge we determined serum ALT levels (B), H&E staining of liver sections (C), hydroxyproline levels in liver homogenate (D), Sirius Red staining of liver sections(E), and immunostaining with α-SMA and myeloperoxidase (MPO) to evaluate neutrophilic infiltration (F) in the different groups. Data are mean±SEM, in B, n=5 animals per group and *p≤ 0.01 vs. corn oil-treated mice; in D, n=5 animals per group and *p≤ 0.01 vs. corn oil-treated mice, and #p≤ 0.05 vs. CCl4+vehicle-treated mice.
DISCUSSION
Cathepsins are lysosomal proteases that have been mainly involved and best characterized in tumorigenesis and metastasis due to extensive ECM remodeling and in the regulation of cell death pathways (18, 21, 45, 46). Although cathepsins have been implicated in liver fibrogenesis as well (22-25), their role in the regulation of HSC biology and their fibrogenic potential is largely unknown. The present report provides evidence for a critical role of cathepsins, particularly CtsB and CtsD, in HSC activation and proliferation, modulating their transdifferentiation into myofibroblasts-like phenotype that ultimately controls liver fibrogenesis.
We show that CtsB and CtsD expression increase during in vitro mouse HSC activation and that this upregulation occurs largely in lysosomes, although a significant fraction is secreted in the extracellular media. The colocalization of cathepsins with markers of early and late endosomes indicates that the release of lysosomal cathepsins follows the secretory pathway to potentially modulate ECM components and plasma membrane HSC receptors.
Previous studies reported that secreted CtsB contributed to the degradation of PDGFβR (41), suggesting that the inability of PDGF to induce HSC proliferation in quiescent HSC was due to the degradation of PDGFβR. However, our findings portrait a different scenario according to which CtsB would directly modulate HSC proliferation by a novel mechanism. Genetic and pharmacological antagonism of CtsB prevents the proliferation of either activated murine HSC cells or the immortalized human LX2 cells. However, rather than regulating the degradation of PDGFβR in response to its ligand PDGF, we observed that the inhibition of CtsB impairs the phosphorylation of AKT by PDGF. Thus, these findings suggest that CtsB positively regulates the PI3K/AKT pathway in response to PDGF. The exact mechanism or intermediate proteins responsible for the decreased activation of AKT observed in the absence of CtsB activity in vitro after PDGF challenge is of utmost importance and merits further investigation.
In addition to preventing HSC proliferation, downregulation of CtsB and CtsD negatively regulates the expression of phenotypic markers of HSC activation such as α-SMA and TGF-β. These findings indicate that cysteine and aspartyl cathepsins directly modulate the early transdifferentiation and perpetuation phase of HSC activation, critical steps for the regulation of their fibrogenic potential and hence liver fibrogenesis. However, while we observed also a decrease in TGF-β expression in primary mouse HSC, TGF-β mRNA levels did not change in LX2 cells probably reflecting that these cells are more differentiated than 5-day old mouse HSC, and that the regulation of TGF-β at this stage of differentiation may be different, as suggested by others (35, 36).
An important finding of this study is the description of active CtsB and CtsD in the extracellular media of HSC, both in primary mouse HSC and in human LX2 cells. This observation is significant since decreased pH may occur at local sites of tissue injury during inflammation, favoring the activation of secreted cathepsins. In fact, secretion of cathepsins correlates positively with invasiveness and metastatic properties of many tumor entities (7, 17, 31), implying that CtsB facilitates invasion by degrading ECM components or by activating other matrix-degrading proteases like the urokinase-type plasminogen activator (47).
Given the role of CtsB/CtsD in orchestrating the proliferating and fibrogenic potential of activated HSC, we next addressed their impact on liver fibrogenesis in vivo. Extending the previous observations of Canbay et al in the BDL model of liver fibrogenesis (28), we analyzed the role of CtsB in CCl4-mediated liver fibrosis. In agreement, we observed that pharmacological inhibition of CtsB prevented the increase in α-SMA upregulation, collagen synthesis and neutrophil infiltration induced by CCl4. In contrast, while CtsB inhibition in BDL-induced liver fibrosis reduced hepatocellular damage which, was shown to contribute to HSC activation (28), we observed that CtsB inhibition did not ameliorate the liver damage caused by CCl4, probably reflecting that the involvement of CtsB in hepatocellular damage may well be stimuli dependent (28, 43, 44). Besides, the upregulation of CtsB during CCl4 administration was confined to HSC, with almost undetectable levels found in hepatocytes, as evidenced by its colocalization with GFAP, and by biochemical analyses of CtsB activity. In this regard and given the nature of GFAP staining, we cannot rule out the participation of CtsB in other myofibroblastic populations in the liver (42, 48). Similarly, and given the complex post-translational regulation of cathepsins (37, 38), we cannot discard the involvement of other related family members in the activation of HSC and modulation of their fibrogenic potential.
Finally, a relevant aspect of our findings is that the CtsB inhibitor, Ca074Me, administered 2 weeks after CCl4 treatment to allow the initial activation of HSC, was able to reduce fibrosis progression. Although further research in this area is needed, our results position HSC cathepsins as potential therapeutic targets for the treatment of liver fibrosis.
Supplementary Material
Acknowledgments
The technical assistance of Susana Nuñez is highly appreciated. The authors thank Dr. Ramón Bataller for generously providing the LX2 cell line, and Drs. C. García-Ruiz, A. Colell and A. Morales for critically reading the manuscript. M.M. is an IDIBAPS Investigator funded by “Department de Salut de la Generalitat de Catalunya”.
Financial support: The work was supported by CIBEREHD and grant PI070193 (Instituto de Salud Carlos III); by grant SAF2006-06780 (Plan Nacional de I+D), Spain; and by grant P50-AA-11999 (Research Center for Liver and Pancreatic Diseases, US National Institute on Alcohol Abuse and Alcoholism).
List of Abbreviations
- BDL
bile duct ligation
- CtsB
cathepsin B
- CtsD
cathepsin D
- ECM
extracellular matrix
- GFAP
glial fibrillary acidic protein
- HSC
hepatic stellate cells
- PDGF
Platelet-derived growth factor
- PDGFβR
PDGF-β receptor
References
- 1.Friedman SL. Hepatic Stellate Cells: Protean, Multifunctional, and Enigmatic Cells of the Liver. Physiol Rev. 2008;88:125–172. doi: 10.1152/physrev.00013.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Tsukada S, Parsons CJ, Rippe RA. Mechanisms of liver fibrosis. Clin Chim Acta. 2006;364:33–60. doi: 10.1016/j.cca.2005.06.014. [DOI] [PubMed] [Google Scholar]
- 3.Iredale JP. Models of liver fibrosis: exploring the dynamic nature of inflammation and repair in a solid organ. J Clin Invest. 2007;117:539–48. doi: 10.1172/JCI30542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bataller R, Brenner DA. Liver fibrosis. J Clin Invest. 2005;115:209–18. doi: 10.1172/JCI24282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kisseleva T, Brenner DA. Hepatic stellate cells and the reversal of fibrosis. J Gastroenterol Hepatol. 2006;21:S84–7. doi: 10.1111/j.1440-1746.2006.04584.x. [DOI] [PubMed] [Google Scholar]
- 6.Bachem MG, Meyer D, Melchior R, M Sell M, Gressner AM. Activation of rat liver perisinusoidal lipocytes by transforming growth factors derived from myofibroblastlike cells. A potential mechanism of self perpetuation in liver fibrogenesis. J Clin Invest. 1992;89:19–27. doi: 10.1172/JCI115561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Joyce JA, Baruch A, Chehade K, Meyer-Morse N, Giraudo E, Tsai FY, et al. Cathepsin cysteine proteases are effectors of invasive growth and angiogenesis during multistage tumorigenesis. Cancer Cell. 2004;5:443–453. doi: 10.1016/s1535-6108(04)00111-4. [DOI] [PubMed] [Google Scholar]
- 8.Liaudet-Coopman E, Beaujouin M, Derocq D, Garcia M, Glondu-Lassis M, Laurent-Matha V, et al. Cathepsin D: newly discovered functions of a long-standing aspartic protease in cancer and apoptosis. Cancer Lett. 2006;237:167–79. doi: 10.1016/j.canlet.2005.06.007. [DOI] [PubMed] [Google Scholar]
- 9.Urbich C, Heeschen C, Aicher A, Sasaki K, Bruhl T, Farhadi MR, et al. Cathepsin is required for endothelial progenitor cell-induced neovascularization. Nat Med. 2005;11:206–213. doi: 10.1038/nm1182. [DOI] [PubMed] [Google Scholar]
- 10.Nakagawa T, Roth W, Wong P, Nelson A, Farr A, Deussing J, et al. Cathepsin L: critical role in Ii degradation and CD4 T cell selection in the thymus. Science. 1998;280:450–3. doi: 10.1126/science.280.5362.450. [DOI] [PubMed] [Google Scholar]
- 11.Saftig P, Hetman M, Schmahl W, Weber K, Heine L, Mossmann H, et al. Mice deficient for the lysosomal proteinase cathepsin D exhibit progressive atrophy of the intestinal mucosa and profound destruction of lymphoid cells. Eur Mol Bio J. 1995;14:3599–3608. doi: 10.1002/j.1460-2075.1995.tb00029.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Nakanishi H, Zhang J, Koike M, et al. Involvement of nitric oxide released from microglia-macrophages in pathological changes of cathepsin D-deficient mice. J Neurosci. 2001;21:7526–7533. doi: 10.1523/JNEUROSCI.21-19-07526.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Koike M, Shibata M, Ohsawa Y, Nishioku T, Okamoto Y, Kominami E, et al. Involvement of two different cell death pathways in retinal atrophy of cathepsin D-deficient mice. Mol Cell Neurosci. 2003;22:146–161. doi: 10.1016/s1044-7431(03)00035-6. [DOI] [PubMed] [Google Scholar]
- 14.Koblinski JE, Ahram M, Sloane BF. Unraveling the role of proteases in cancer. Clin Chim Acta. 2000;291:113–135. doi: 10.1016/s0009-8981(99)00224-7. [DOI] [PubMed] [Google Scholar]
- 15.Laurent-Matha V, Maruani-Herrmann S, Prébois C, Beaujouin M, Glondu M, Noël A, et al. Catalytically-inactive human cathepsin D triggers fibroblast invasive growth. J Cell Biol. 2005;168:489–499. doi: 10.1083/jcb.200403078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Fehrenbacher N, Jaattela M. Lysosomes as targets for cancer therapy. Cancer Res. 2005;65:2993–2995. doi: 10.1158/0008-5472.CAN-05-0476. [DOI] [PubMed] [Google Scholar]
- 17.Khalkhali-Ellis Z, Hendrix MJ. Elucidating the function of secreted maspin: inhibiting cathepsin D-mediated matrix degradation. Cancer Res. 2007;67:3535–3539. doi: 10.1158/0008-5472.CAN-06-4767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Conus S, Perozzo R, Reinheckel T, Peters C, Scapozza L, Yousefi S, et al. Caspase-8 is activated by cathepsin D initiating neutrophil apoptosis during the resolution of inflammation. J Exp Med. 2008;205:685–698. doi: 10.1084/jem.20072152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Rochefort H, Liaudet-Coopman E. Cathepsin D in cancer metastasis: a protease and a ligand. APMIS. 1999;107:86–95. doi: 10.1111/j.1699-0463.1999.tb01530.x. [DOI] [PubMed] [Google Scholar]
- 20.Turk V, Kos J, Turk B. Cysteine cathepsins (proteases)--on the main stage of cancer? Cancer Cell. 2004;5:409–410. doi: 10.1016/s1535-6108(04)00117-5. [DOI] [PubMed] [Google Scholar]
- 21.Mohamed MM, Sloane BF. Cysteine cathepsins: multifunctional enzymes in cancer. Nat Rev Cancer. 2006;6:764–75. doi: 10.1038/nrc1949. [DOI] [PubMed] [Google Scholar]
- 22.Gohda E, Nagahama J, Nakamura O, Tsubouchi H, Daikuhara Y, Pitot HC. Increased activities of liver cathepsins T and D in carbon tetrachloride-treated rats. Biochim Biophys Acta. 1984;802:362–371. doi: 10.1016/0304-4165(84)90184-3. [DOI] [PubMed] [Google Scholar]
- 23.Yamamoto H, Murawaki Y, Kawasaki H. Collagenolytic cathepsin B and L activity in experimental fibrotic liver and human liver. Res Commun Chem Pathol Pharmacol. 1992;76:95–112. [PubMed] [Google Scholar]
- 24.Leto G, Tumminello FM, Pizzolanti G, Montalto G, Soresi M, Gebbia N. Lysosomal cathepsins B and L and stefin A blood levels in patients with hepatocellular carcinoma and/or liver cirrhosis: potential clinical implications. Oncology. 1997;54:79–83. doi: 10.1159/000227666. [DOI] [PubMed] [Google Scholar]
- 25.Leto G, Tumminello FM, Pizzolanti G, Montalto G, Soresi M, Ruggeri I, et al. Cathepsin D serum mass concentrations in patients with hepatocellular carcinoma and/or liver cirrhosis. Eur J Clin Chem. 1996;34:555–560. doi: 10.1515/cclm.1996.34.7.555. [DOI] [PubMed] [Google Scholar]
- 26.Kristensen DB, Kawada N, Imamura K, Miyamoto Y, Tateno C, Seki S, et al. Proteome analysis of rat hepatic stellate cells. Hepatology. 2000;32:268–277. doi: 10.1053/jhep.2000.9322. [DOI] [PubMed] [Google Scholar]
- 27.Takashima T, Kawada N, Maeda N, Okuyama H, Uyama N, Seki S, et al. Pepstatin A attenuates the inhibitory effect of N-acetyl-L-cysteine on proliferation of hepatic myofibroblasts (stellate cells) Eur J Pharmacol. 2002;451:265–270. doi: 10.1016/s0014-2999(02)02296-3. [DOI] [PubMed] [Google Scholar]
- 28.Canbay A, Guicciardi ME, Higuchi H, Feldstein A, Bronk SF, Rydzewski R, et al. Cathepsin B inactivation attenuates hepatic injury and fibrosis during cholestasis. J Clin Invest. 2003;112:152–159. doi: 10.1172/JCI17740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kim JS, Shukla SD. Histone h3 modifications in rat hepatic stellate cells by ethanol. Alcohol Alcohol. 2005;40:367–372. doi: 10.1093/alcalc/agh170. [DOI] [PubMed] [Google Scholar]
- 30.Barrett J. Fluorimetric assays for cathepsin B and cathepsin H with Methylcoumarylamide Substrates. Biochem J. 1980;187:909–912. doi: 10.1042/bj1870909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Klose A, Wilbrand-Hennes A, Zigrino P, Weber E, Krieg T, Mauch C, et al. Contact of high-invasive, but not low-invasive, melanoma cells to native collagen I induces the release of mature cathepsin B. Int J Cancer. 2006;118:2735–2743. doi: 10.1002/ijc.21700. [DOI] [PubMed] [Google Scholar]
- 32.Wang X, Tang X, Gong X, Albanis E, Friedman SL, Mao Z. Regulation of hepatic stellate cell activation and growth by transcription factor myocyte enhancer factor 2. Gastroenterology. 2004;127:1174–1788. doi: 10.1053/j.gastro.2004.07.007. [DOI] [PubMed] [Google Scholar]
- 33.Lee M, Fridman R, Mobashery S. Extracellular proteases as targets for treatment of cancer metastases. Chem Soc Rev. 2004;33:401–409. doi: 10.1039/b209224g. [DOI] [PubMed] [Google Scholar]
- 34.Leto G, Tumminello FM, Gebbia N, Rausa L. Kinetics of in vivo inhibition of tissue cathepsin D by pepstatin A. Int J Biochem. 1988;20:917–920. doi: 10.1016/0020-711x(88)90175-9. [DOI] [PubMed] [Google Scholar]
- 35.Xu L, Hui AY, Albanis E, Arthur MJ, O’Byrne SM, Blaner WS, et al. Human hepatic stellate cell lines, LX-1 and LX-2: new tools for analysis of hepatic fibrosis. Gut. 2005;54:142–151. doi: 10.1136/gut.2004.042127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Liu C, Gaça MD, Swenson ES, Vellucci VF, Reiss M, Wells RG. Smads 2 and 3 are differentially activated by transforming growth factor-beta (TGF-beta) in quiescent and activated hepatic stellate cells. Constitutive nuclear localization of Smads in activated cells is TGF-beta-independent. J Biol Chem. 2003;278:11721–11728. doi: 10.1074/jbc.M207728200. [DOI] [PubMed] [Google Scholar]
- 37.Dooley S, Delvoux B, Streckert M, Bonzel L, Stopa M, ten Dijke P, et al. Transforming growth factor beta signal transduction in hepatic stellate cells via Smad2/3 phosphorylation, a pathway that is abrogated during in vitro progression to myofibroblasts. TGFbeta signal transduction during transdifferentiation of hepatic stellate cells. FEBS Lett. 2001;502:4–10. doi: 10.1016/s0014-5793(01)02656-4. [DOI] [PubMed] [Google Scholar]
- 38.Turk V, Turk B, Guncar G, Turk D, Kos J. Lysosomal cathepsins: structure, role in antigen processing and presentation, and cancer. Adv Enzyme Regul. 2002;42:285–303. doi: 10.1016/s0065-2571(01)00034-6. [DOI] [PubMed] [Google Scholar]
- 39.Laurent-Matha V, Derocq D, Prébois C, Katunuma N, Liaudet-Coopman F. Processing of human Cathepsin D is independent of its catalytic function and auto-activation: Involvement of cathepsins L and B. J Biochem. 2006;139:363–371. doi: 10.1093/jb/mvj037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Nishimura Y, Kawabata T, Kato K. Identification of latent procathepsins B and L in microsomal lumen: Characterization of enzymatic activation and proteolytic processing in vitro. Arch Biochem Biophys. 1988;261:64–71. doi: 10.1016/0003-9861(88)90104-x. [DOI] [PubMed] [Google Scholar]
- 41.Okuyama H, Shimahara Y, Kawada N, Seki S, Kristensen DB, Yoshizato K, et al. Regulation of cell growth by Redox-mediated Extracellular proteolysis of Platelet-derived Growth Factor Receptor β. J Biol Chem. 2001;30:28274–28280. doi: 10.1074/jbc.M102995200. [DOI] [PubMed] [Google Scholar]
- 42.Geerts A. History, Heterogeneity, Developmental Biology, and Functions of Quiescent Hepatic Stellate Cells. Semin Liver Dis. 2001;21:311–335. doi: 10.1055/s-2001-17550. [DOI] [PubMed] [Google Scholar]
- 43.Guicciardi ME, Deussing J, Miyoshi H, Bronk SF, Svingen PA, Peters C, et al. Cathepsin B contributes to TNF-alpha-mediated hepatocyte apoptosis by promoting mitochondrial release of cytochrome c. J Clin Invest. 2000;106:1127–37. doi: 10.1172/JCI9914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Li Z, Berk M, McIntyre TM, Gores GJ, Feldstein AE. The lysosomal-mitochondrial axis in free fatty acid-induced hepatic lipotoxicity. Hepatology. 2008;47:1495–1503. doi: 10.1002/hep.22183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Heinrich M, Wickel M, Schneider-Brachert W, Sandberg C, Gahr J, Schwandner R, et al. Cathepsin D targeted by acid sphingomyelinase-derived ceramide. EMBO J. 1999;18:5252–5263. doi: 10.1093/emboj/18.19.5252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Heinrich M, Neumeyer J, Jakob M, Hallas C, Tchikov V, Winoto-Morbach S, et al. Cathepsin D links TNF-induced acid sphingomyelinase to Bid-mediated caspase-9 and -3 activation. Cell Death Differ. 2004;11:550–563. doi: 10.1038/sj.cdd.4401382. [DOI] [PubMed] [Google Scholar]
- 47.Krueger S, Haeckel C, Buehling F, Roessner A. Inhibitory effects of antisense cathepsin B cDNA transfection on invasion and motility in a human osteosarcoma cell line. Cancer Res. 1999;59:6010–6014. [PubMed] [Google Scholar]
- 48.Knittel T, Kobold D, Piscaglia F, Saile B, Neubauer K, Mehde M, et al. Localization of liver myofibroblasts and hepatic stellate cells in normal and diseased rat livers: distinct roles of (myo)fibroblast subpopulations in hepatic tissue repair. Histochem Cell Biol. 1999;112:387–401. doi: 10.1007/s004180050421. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.