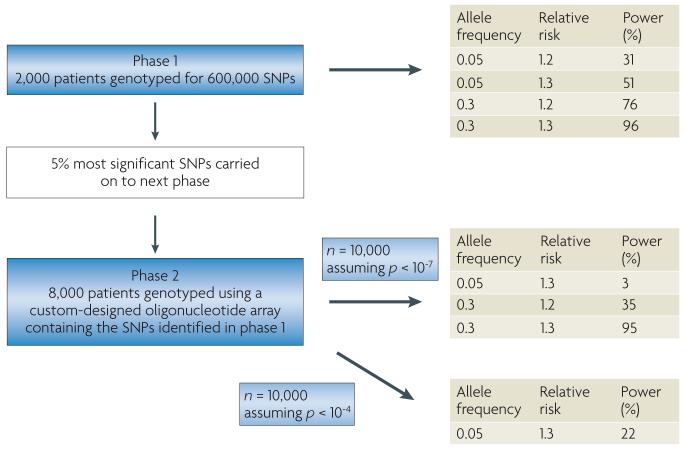
Figure 3. Proposed design for a radiation toxicity genome-wide association study (GWAS).
In the first phase of a staged approach a full set of tagged single nucleotide polymorphisms (tag-SNPs; 600,000) is chosen that comprehensively captures common variations across the genome. This set is genotyped using a whole genome chip in a relatively small population and at a liberal p-value threshold, to identify a subset of SNPs with putative associations. As the phenotype of interest exhibits a range of responses, including intermediate levels, this will give greater power than a case–control study of the same size. Phase II takes the top 5% of SNPs identified in phase I or those passing an initial threshold (or filter) and re-tests these SNPs using a custom-designed oligonucleotide array in a larger independent population sample, thus significantly increasing efficiency and reducing genotyping costs.