Abstract
Retinoid-related orphan nuclear receptors (RORs) α and γ (NR1F1, -3) are highly expressed in liver, adipose tissue, thymus, and brain and are involved in many physiological processes, such as circadian rhythm and immune function. Enzymes in the cytochrome P450 2C subfamily metabolize many clinically important drugs and endogenous compounds, such as the anticancer drug paclitaxel and arachidonic acid, and are highly expressed in liver. Here, we present the first evidence that RORs regulate the transcription of human CYP2C8. Overexpression of RORα and RORγ in HepG2 cells significantly enhanced the activity of the CYP2C8 promoter but not that of the CYP2C9 or CYP2C19 promoters. Computer analyses, promoter deletion studies, gel shift assays, and mutational analysis identified an essential ROR-responsive element at -2045 base pairs in the CYP2C8 promoter that mediates ROR transactivation. Adenoviral overexpression of RORα and -γ significantly induced endogenous CYP2C8 transcripts in both HepG2 cells and human primary hepatocytes. Knockdown of endogenous RORα and -γ expression in HepG2 cells by RNA interference decreased the expression of endogenous CYP2C8 mRNA by ∼50%. These data indicate that RORs transcriptionally up-regulate CYP2C8 in human liver and, therefore, may be important modulators of the metabolism of drugs and physiologically active endogenous compounds by this enzyme in liver and possibly extrahepatic tissues where RORs are expressed.
The human CYP2C subfamily consists of four genes: CYP2C8, CYP2C9, CYP2C18, and CYP2C19. CYP2C18 does not seem to be expressed at the protein level. The other three CYP2C enzymes metabolize approximately 20% of currently used clinical drugs, including the antidiabetic drugs tolbutamide and rosiglitazone, the anticoagulant drug warfarin, the anticonvulsant phenytoin, the antiulcer drug omeprazole, the anticancer drug paclitaxel, and numerous nonsteroidal anti-inflammatory drugs, such as ibuprofen (Goldstein, 2001). They are also involved in the oxidative metabolism of endogenous compounds, such as arachidonic acid and retinoic acid.
It is well known that there is marked variability in the metabolism of CYP2C substrates in human populations because of the occurrence of genetic polymorphisms in the CYP2C genes. The expression of CYP2C enzymes also has been reported to be induced by various drugs, including rifampicin, hyperforin (the active constituent in St. John's Wort), phenobarbital, and dexamethasone (Raucy et al., 2002; Madan et al., 2003; Komoroski et al., 2004). This induction further amplifies the individual variability in drug metabolism in human populations and may lead to a change in the half-life of drugs and then eventually result in drug tolerance or therapeutic failure.
A number of nuclear receptors have been discovered to play important roles in the mediation of transactivation of P450 enzymes. In many cases, these nuclear receptors bind ligands, which may be involved in initiating translocation of the receptor to the nucleus, where the receptor binds to responsive elements within gene promoters and recruits coactivators to affect chromatin structure and increase the transcription of target genes (Handschin and Meyer, 2003). Studies from our laboratory and other laboratories have revealed that several nuclear receptors that mediate drug-induced transactivation of CYP2C9,-2C8, and -2C19 genes include the constitutive androstane receptor (CAR), the pregnane X receptor (PXR), and the glucocorticoid receptor (Gerbal-Chaloin et al., 2002; Chen et al., 2003, 2004; Ferguson et al., 2005). On the other hand, the constitutive regulation of the CYP2C genes is believed to be regulated in liver by receptors that include the hepatic nuclear factor (HNF) 4α, HNF3γ, and CCAAT/enhancer-binding protein (Jover et al., 1998, 2001; Bort et al., 2004; Chen et al., 2005). HNF4α also seems to cross-talk with CAR/PXR, which binds to more distal CAR/PXR-responsive elements to achieve synergistic activation with CAR and increase PXR-mediated induction by rifampicin (Chen et al., 2005).
Another family of receptors, the retinoic acid receptor-related orphan receptors, consists of three members, RORα, -β, and -γ (Jetten and Joo, 2006). Several isoforms with varying lengths of the N terminus occur because of alternative promoter usage. They bind as a monomer to ROR-responsive elements (ROREs) consisting of a 6-bp AT-rich sequence preceding the half-core PuGGTCA motif and act as either transcription activators or repressors, depending on which cofactors are recruited. Studies from ROR knockout mice have revealed that these receptors exhibit critical roles in a number of physiological and pathological processes, including cerebellar ataxia, inflammation, atherosclerosis, immune response, and circadian rhythm (Jetten and Joo, 2006). Recent studies have shown that the levels of several phase I and II enzymes, including Cyp2c70, Cyp2c39, and Cyp7b1, were altered in livers of RORα and RORγ knockout mice, indicating that RORs may be a new regulator for P450 enzymes (Kang et al., 2007). Wada et al. (2008) further demonstrated that RORα could bind to the murine Cyp7b1 promoter to directly transactivate this gene.
In humans, RORα1, -α4, and -γ1 are the forms expressed in liver, but only a few hepatic genes, including apolipoprotein C-III (RORα1) (Jetten and Joo, 2006), apolipoprotein A5 (RORα1 and -α4) (Genoux et al., 2005), and hepatic synthesis of the plasma protein B-fibrinogen (Chauvet et al., 2005) have been identified as RORα1 and -α4 targets. Because recent knockout studies indicated several murine P450 genes may be targets for ROR regulation in liver (Kang et al., 2007), we hypothesized that RORs might also be transcriptional modulators of human P450 genes, contributing to the regulation of gene expression in liver and various extrahepatic tissues. In this study, we investigated the regulation of human CYP2C8,-2C9, and -2C19 genes by RORα1, RORα4, and RORγ1. We provide experimental evidence showing that both RORα and RORγ up-regulate the transcription of CYP2C8 in HepG2 cells, human primary hepatocytes, and Caco-2 cells by activating the CYP2C8 promoter directly through binding an RORE in the promoter region. The observations suggest an important role for RORs in the regulation of CYP2C8 expression in human liver and, as a consequence, a role in drug metabolism.
Materials and Methods
Promoter Constructs and Expression Plasmids. The wild-type CYP2C9-2923, CYP2C8-2966, CYP2C8-2527, and CYP2C19-2.7k/pGL3_Basic constructs were as described previously (Chen et al., 2003; Ferguson et al., 2005). Of the CYP2C8 promoter region, -2 and -1.5 kb were amplified by PCR and constructed into pGL3_Basic to yield 2C8-2k and -1.5k promoter constructs (Ferguson et al., 2005). CYP2C8-2966 was used as the template to produce two mutants (CYP2C8-2966/-2289m and CYP2C8-2966/-2045m) by using QuikChange Site-directed mutagenesis (Stratagene, La Jolla, CA) and a deletion construct, CYP2C8-2966/ΔBglII, by BglII digestion. The forward primers utilized for mutagenesis are as follows (the hexamer half-sites are indicated by boldface capital letters and mutated nucleotides are underlined): distal RORE mutation, 5′ cttttattctataaAAAGAAACCTCAaggcagg 3′; and proximal RORE-mutation, 5′ ccaaacacgtcTGAGGCACATTTtactc 3′. DNA sequencing was performed for all constructs to verify the mutations and to assure that no spurious mutations occurred.
The cDNAs of mouse RORα4 (Y08640), RORγ1 (NM_011281), RORγ2 (AF163668), and human RORα1 (NM_134261) were prepared by RT-PCR and inserted into p3XFLAG-CMV.7.1 (Sigma-Aldrich, St. Louis, MO) to produce the wild-type expression plasmids for transient transfection. We used murine RORα4, -γ1, and -γ2 expression plasmids for these studies because the amino acid identity between human and murine RORs is as high as 97% for RORα proteins and 89% for RORγ proteins, and their biological effects are very similar. Moreover, we had constructed several DNA binding mutants and AF2 deletion mutants of the murine RORα and -γ. The wild-type murine ROR expression constructs were used as the templates to produce three mutants: RORα4C13A, which has no DNA binding ability; RORγ1ΔAF2 [deletion of Pro496 through the C terminus (20 amino acids including the AF2 domain, PPLYKELFSTDVESPEGLSK)]; and RORγ2ΔAF2, which has the same deletion as RORγ1ΔAF2. To generate the ROR expression plasmids for in vitro transcription and translation, the cDNA of mouse RORα4 was cloned into pTRIamp18 (Ambion, Austin, TX), whereas the cDNAs of mouse RORγ1 and human RORα1 were constructed into pcDNA3.1 (Invitrogen, Carlsbad, CA).
Cell Culture and Transfection. HepG2 and Caco-2 cells were maintained in minimum essential medium supplemented with 10 to ∼20% fetal bovine serum, 2 mM l-glutamine, 1 mM sodium pyruvate, and antibiotics at 37°C and under 5% CO2. For transient transfection, cells were seeded into a 24-well plate at a density of 105/well the day before transfection. To examine the activation of CYP2C8 promoters by RORs, an Effectene transfection kit (QIA-GEN, Valencia, CA) was used to complete transient transfections in HepG2 cells with CYP2C promoter constructs and pGL3_Basic, along with the internal control pRL-thymidine kinase promoter. Expression constructs for nuclear receptors or empty expression plasmids were cotransfected in parallel. The total amount of transfected plasmids was 200 ng/transfection. Twenty-four hours later, medium was changed, and cells were maintained for another day, followed by dual luciferase assays (Promega, Madison, WI). Firefly luciferase activities were normalized to Renilla luciferase readings to calculate relative promoter activity. Analogous procedures were used for the examination of other deleted and mutated promoter constructs.
Gel Shift Assays. RORα4, -α1, and -γ1 were synthesized in vitro using the TNT T7 Quick Coupled Transcription Translation System (Promega), following the manufacturer's protocol. The empty expression vector was used as control. Gel shift assays were performed essentially as described previously (Medvedev et al., 1996). To decrease the nonspecific binding, 1 μg of sonicated salmon sperm DNA and 1 μg of single-strand DNA were added to the incubation mixture. The following oligonucleotides were used as probes, wild-type, or mutated specific cold competitors. The hexamer half-sites are indicated by boldface capital letters, and mutated nucleotides are underlined: CYP2C8-2289RORE, 5′-ctagTAAAAAGAAAGGTCAAGG-3′; CYP2C8-2289ROREmut, 5′-ctagTAAAAAGAAACCTCAAGG-3′; CYP2C8-2045RORE, 5′-ctagGTCTGACCCACATTTTAC-3′; CYP2C8-2045ROREmut, 5′-ctagGTCTGAGGCACATTTTAC-3′; CYP2C8-2045ROREmut2, 5′ ctagGTCTGACCCACGTTTTAC-3′; and mouse Purkinje cell protein-2, 5′-ctagGTTATAGTAACTGGGTCAGGGGACTC-3′.
Adenovirus Preparation and Infection. cDNA of FLAG-mRORα4 (murine RORα4) and FLAG-mRORγ1 (murine RORγ1) were used to prepare ROR adenoviruses for overexpression in HepG2 cells by using AdEasy-technology (Qbiogene Inc., Irvine, CA). Adenoviral infection was performed in HepG2 cells with adenoviruses LacZ and RORα4 or GFP/β-Gal and RORγ1 (1000 particles/cell) for 1 to 4 days, followed by total RNA isolation with an RNeasy Mini kit (QIAGEN). Human primary hepatocytes purchased from CellzDirect/Invitrogen (Carlsbad, CA) were cultured in Williams' medium E (Sigma-Aldrich) complemented with 1× ITS+ supplement (Sigma-Aldrich; containing 10 μg/ml insulin from bovine pancreas, 5.5 μg/ml human transferrin, 5 ng/ml sodium selenite, 0.5 mg/ml bovine serum albumin, and 4.7 μg/ml linoleic acid), 15 mM HEPES, 2 mM l-glutamine, 10 nM dexamethasone, and penicillin (50 U/ml)/streptomycin (50 μg/ml). Hepatocytes were infected by viruses for 60 h, and total RNA was isolated for quantitative RT-PCR to measure mRNA concentration for CYP2C8, murine and human RORα, murine and human RORγ, and TBP as the endogenous control.
Quantitative RT-PCR. Total RNA was extracted using an RNeasy Mini prep system (QIAGEN). RT-PCR analysis was performed in two steps by initial reaction with Moloney murine leukemia virus reverse transcriptase (Applied Biosystems, Foster City, CA). PCR with Taqman Universal PCR Master Mix (Applied Biosystems) was then performed with gene-specific primers using relative quantification methods (2-ΔCt) and measured on an Applied Biosystems 7900HT Sequence Detection System using Taqman probes for CYP2C8, RORα, RORγ, HNF4α, GAPDH, mRORα, mRORγ, and the endogenous control TBP.
Western Blot Analysis. To analyze the exogenous ROR protein expression in HepG2 cells, whole-cell lysates were prepared from HepG2 cells infected with control GFP virus or adenoviruses containing the different FLAG-tagged RORs in six-well plates by using radioimmunoprecipitation assay buffer (Promega). Equal volumes of lysates were subjected to 4 to 12% NuPAGE gel electrophoresis (Invitrogen), transferred to nitrocellulose membranes, and blocked with 5% nonfat milk blocking buffer. Horseradish peroxidase-conjugated monoclonal anti-FLAG antibody (Sigma-Aldrich) was used for immunoblotting at room temperature for 1 h, and then the membrane was washed six times. Detection was achieved using a Super Signal West Femto kit (Pierce Chemical, Rockford, IL).
Reduction of Endogenous Gene Expression by Small Interference RNAs. To reduce endogenous ROR expression in HepG2 cells, cells were seeded into a 24-well plate (105/well) and transfected the next day with small interference RNAs (siRNAs) against human RORα and RORγ (Dharmacon RNA Technologies, Lafayette, CO) at two doses of 50 and 100 nM following the instructions for Dharma-FECT 4 transfection reagent. Nonspecific siRNA and siRNA against GAPDH were transfected at the same time as the negative and positive controls, respectively. Twenty-four hours after transfection, cells were harvested, and total RNAs were isolated. Quantitative RT-PCR was performed to analyze mRNA levels of human RORα, RORγ, GAPDH, and CYP2C8.
Statistical Analysis. Statistical analysis was performed in SigmaStat version 3.5 (Systat Software, Inc., San Jose, CA) using either one- or two-way analysis of variance as appropriate; subsequent pair-wise comparisons were made using the Bonferroni t-test method.
Results
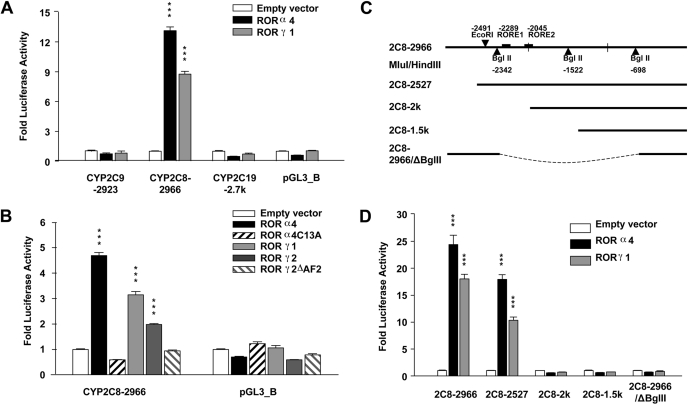
RORs Specifically Transactivate the CYP2C8 Promoter in HepG2 Cells. To examine whether RORs can activate the transcription of human CYP2C genes, expression plasmids for the murine nuclear receptors RORα4 and RORγ1 were cotransfected into HepG2 cells along with the luciferase promoter constructs CYP2C9-2923, CYP2C8-2966, CYP2C19-2.7k, or the empty luciferase vector pGL3_Basic. Among the CYP2C promoters, only the CYP2C8-2966 promoter was strongly activated by RORα4 and RORγ1 (13- and 9-fold, respectively, p < 0.001), whereas the CYP2C9 and CYP2C19 promoters showed no response to exogenous RORs (Fig. 1A). The nonresponsiveness of the CYP2C9 and CYP2C19 promoters to ROR was confirmed using 12-kb upstream promoter constructs for these two genes in transfection assays in HepG2 cells (data not shown). To confirm the specificity of activation of the CYP2C8 promoter by RORs, we cotransfected HepG2 cells with CYP2C8 promoter constructs and expression plasmids containing two ROR mutants and wild-type RORα4, RORγ1, and RORγ2. The mutant RORα4C13A harbors a mutation at its DNA binding domain and does not bind to ROR-responsive elements in DNA, whereas in RORγ2ΔAF2, the AF2 domain is truncated and, therefore, unable to recruit coactivators. As shown in Fig. 1B, wild-type RORα4, RORγ1, and RORγ2 significantly elevated the activity of CYP2C8 promoter (p < 0.001), whereas the two mutants had no significant effect on promoter activity. These data further confirm that the CYP2C8 promoter is transactivated by RORα4, RORγ1, and RORγ2. We then examined the effect of progressive deletions of the CYP2C8 promoter construct (Fig. 1C) on ROR-mediated transactivation to identify the region in the CYP2C8 promoter that mediates ROR activation. As shown in Fig. 1D, significant ROR activation of CYP2C8 was observed with the CYP2C8-2966 and CYP2C8-2527 constructs (p < 0.001), but not with CYP2C8-2k and -1.5k constructs, suggesting that ROREs within the 500-bp fragment from -2.5 to -2 kb of the CYP2C8 promoter are responsible for this transactivation. We also created a construct in which the distal and proximal promoter regions were retained, but the middle region including this 500-bp fragment was deleted; no ROR activation was observed with the deletion construct. This result further supports the hypothesis that the localization of the functional RORE(s) is within the 500-bp promoter fragment identified above.
Fig. 1.
RORα4 and -γ1 transactivate the CYP2C8 promoter but not the CYP2C9 or CYP2C19 promoters in HepG2 cells. A and B, promoter constructs in pGL3_Basic driven by CYP2C9,-2C8, and -2C19 promoters and empty vector were cotransfected into HepG2 cells along with the internal control pRL-thymidine kinase promoter. Expression vector containing the murine nuclear receptors RORα4, -γ1, -γ2, or their corresponding DNA binding mutant (RORα4C13A) and RORγ2ΔAF2 (lacking the AF2 domain) were cotransfected into cells concomitantly. Twenty-four hours after transfections, cells were refreshed and maintained for another day, followed by cell lysis and luciferase activity assays. C and D, deletion constructs of CYP2C8 were similarly transfected with RORα4 or -γ1. RORα4, -γ1, or -γ2 increased CYP2C8 promoter activity compared with the empty vector (***, p < 0.001). Neither of the ROR mutants activated CYP2C8 promoter activity. Values represent the means of three independent transfections ± S.E.
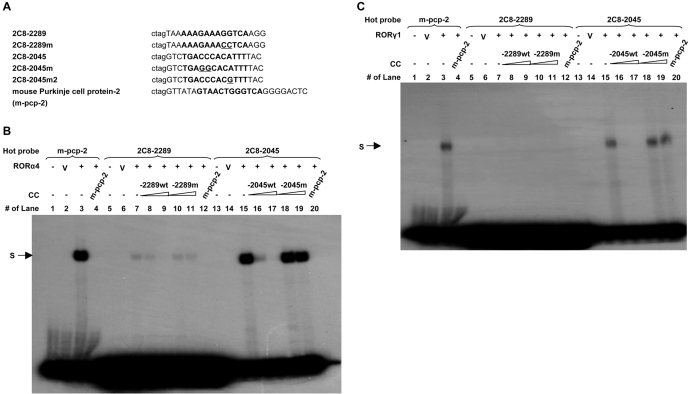
Identification of Two Putative ROREs That Are Required for ROR Activation of the CYP2C8 Promoter. With the sequence of the core motif for RORE, (A/T)6-AGGTCA, we performed a computer search (Motif/Vector NTI) for putative ROREs within the 3 kb of the CYP2C8 promoter and found two putative ROREs at -2289 and -2045 bp, respectively. Both sites are localized within the 500-bp promoter region identified above. Therefore, gel shift assays were performed to determine whether these two putative ROREs bind ROR in vitro. As displayed in Fig. 2B, transcribed and translated in vitro RORα4 products formed strong complexes with the 32P-labeled oligonucleotide probe CYP2C8-2045RORE (lane 15) and with the positive control, a known RORE previously characterized from the murine Purkinje cell protein-2 (m-pcp-2) gene (lane 3), whereas products transcribed and translated in vitro from the empty pCR3.1 vector (V) did not produce any specific bands (lanes 2, 6, and 14 for all radiolabeled probes). In vitro-synthesized RORα4 exhibited a much weaker affinity for the 2C8-2289RORE, despite the fact that 10× reactive probe was added to the gel (Fig. 2B, lane 7). Preincubation with 5× or 50× excess of wild-type cold competitor ROREs effectively competed out the formation of all of these complexes (see lane 4 for m-pcp-2; lanes 8, 9, and 12 for 2C8-2289RORE; and lanes 16, 17, and 20 for 2C8-2045), but mutated cold ROREs were unable to compete for these complexes (lanes 10 and 11 for 2C8-2289RORE, lanes 18 and 19 for 2C8-2045RORE). In vitro-transcribed RORγ1 also bound both the positive control m-pcp-2 probe and the 2C8-2045RORE (Fig. 2C, lanes 3 and 15). Again, the wild-type cold probes could effectively compete out the formation of these complexes (see lane 4 for m-pcp-2 and lanes 16, 17 and 20 for 2C8-2045RORE), whereas mutated ROREs did not (lanes 18 and 19), showing the specificity of these complexes. We could not detect complex formation between the 2C8-2289RORE and the in vitro-transcribed RORγ1.
Fig. 2.
Identification of the -2289 and -2045RORE within the CYP2C8 promoter by gel shift assays. A, sequences of the oligonucleotides used for the binding assay for Figs. 2 and 3. Known or putative ROREs are written in bold and capital, the point mutations are underlined, and the nucleotides in lower case were added for labeling. B and C, 32P-labeled probes for two putative ROREs within CYP2C8 (at -2289 and -2045 bp, respectively) and one control known RORE from m-pcp-2 were incubated at room temperature for 20 min with (+) or without (-) in vitro-transcribed and -translated ROR proteins RORα4 (B) and RORγ1 (C). Lanes 2, 6, and 14 (V) contained an in vitro translation transcription reaction carried out with empty vector rather than one carried out with vector containing an ROR. For competition analysis, 5× or 50× excess of various cold competitors (CCs) was added into binding reactions. The RORE at -2045 bp in the CYP2C8 promoter bound RORs much more strongly than the element at -2289 bp (B and C). Wild-type but not mutated cold competitors effectively competed for both elements showing the specificity of binding.
To further examine the binding of RORs to the 2C8-2045RORE, antibody against RORα and goat IgG was included in the incubation mixture before the addition of hot probes. A supershifted complex with retarded mobility was observed with the specific RORα antibody, as shown by the arrow (Fig. 3A, lane 16), but not with control IgG (lane 15). We also saw a supershift with ROR antibody to the positive mouse pcp control (Fig. 3A, lane 6). These results demonstrate the existence of RORα in these complexes. Introduction of one mutation in the 2C8-2045RORE oligo (2C8-2045m2) (shown in Fig. 2A) resulted in severe impairment in its ability to compete for the binding to RORs (Fig. 3A, lanes 13 and 14 for RORα4 and lanes 22 and 23 for RORγ1). Another splicing variant, RORα1, is known to be expressed well in human liver and HepG2 cells (Chauvet et al., 2002). In Fig. 3B, gel shift assays clearly show that RORα1 could also bind to both ROREs within the CYP2C8 promoter, although the proximal site again showed higher affinity. The complex could be supershifted by antibody to RORα. Taken together, these data clearly show that both ROREs can interact with RORs but that the 2C8-2045RORE has a much stronger affinity for both RORα and RORγ than the 2C8-2289RORE.
Fig. 3.
Binding of RORα1 and RORα4 to the -2289 and -2045 ROREs within the CYP2C8 promoter by gel shift assays and supershifts with RORα antibody. 32P-labeled probes for the two CYP2C8 ROREs and the control RORE (m-pcp-2) were incubated at room temperature for 20 min with (+) or without (-) in vitro-transcribed and -translated ROR proteins (A, RORα4 and RORγ1; B, RORα1) as described in Fig. 2. Wild-type but not mutated cold competitors effectively competed for both elements confirming the specificity of the binding. Antibody against RORα caused a supershift (SS) of the RORα4 complex with both m-pcp-2 (A, lane 6) and the -2045 RE (A, lane 16), whereas IgG did not (lanes 5 and 15). The supershifts were also formed with the complex with RORα1 by RORα antibody (B, lanes 9 and 18). The RORE at -2045 bp in the CYP2C8 promoter bound RORα1 much more strongly than the element at -2289 bp.
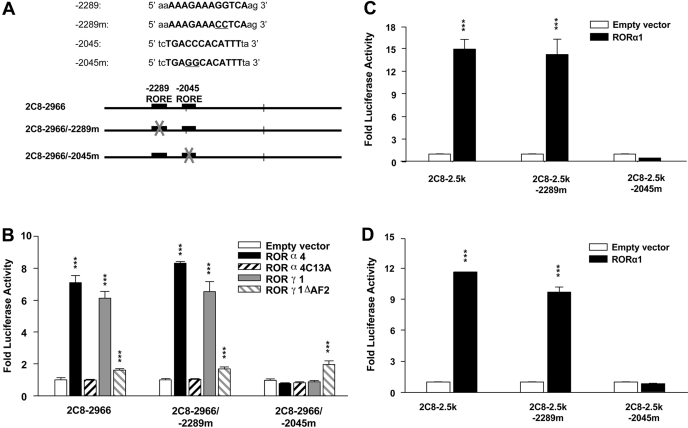
To functionally investigate the roles of these two putative ROREs in the activation of CYP2C8 by RORs, we analyzed the effect of mutations within the two putative ROREs (Fig. 4A) on ROR-mediated activation of the CYP2C8 promoter in HepG2 cells by transient transfection assays. As shown in Fig. 4B, mutation of the -2289RORE did not significantly affect the activation of CYP2C8, either by murine RORα4 or -γ1, whereas mutation of the -2045RORE almost totally abolished the transactivation of the CYP2C8 promoter by RORα4 and RORγ1. In contrast to wild-type ROR, the RORα4 mutant did not significantly increase the activity of the CYP2C8 promoter (p < 0.01), whereas the RORγ1 mutant had much less activity than wild-type RORγ1. Because RORα1 selectively transactivates the ApoIII promoter in human colon carcinoma Caco-2 cells (but not in HepG2 cells), we compared the transactivation of the CYP2C8 promoter by human RORα1 in HepG2 cells and Caco-2 cells. As shown in Fig. 4, C and D, the activity of the CYP2C8 promoter was increased similarly by RORα1 (p < 0.001) in both cell lines, and the mutation of the proximal RORE, but not the distal one, completely abolished this activation. Similar activation of the CYP2C8 promoter by RORα4 and RORγ1 was also observed in Caco-2 cells (data not shown). These data are consistent with those from electrophoretic mobility shift assay and further indicate that the activation of the CYP2C8 promoter by RORs in HepG2 cells and Caco-2 cells is mediated by an interaction of RORs, with an essential RORE at -2045 bp.
Fig. 4.
Mutational analysis of the human CYP2C8 promoter constructs to functionally characterize the two putative ROREs at -2289 and -2045 bp. A, schematic representations of the wild-type and the RORE mutated promoter constructs for transfection. B, HepG2 cells were transfected by wild-type CYP2C8-2966-bp promoter constructs or one of two CYP2C8 promoter constructs containing mutations in ROREs at -2289 or -2045, respectively. Expression plasmids of wild-type murine RORα4, its DNA binding mutant RORα4C13A, wild-type RORγ1, or a mutant lacking the AF2 domain (RORγ1ΔAF2), were cotransfected concomitantly. HepG2 (C) or Caco-2 (D) cells were similarly transfected with normal CYP2C8 promoter constructs or constructs containing mutations at either RORE and expression plasmid containing wild-type human RORα1. Twenty-four hours after transfections, cells were refreshed and maintained for another day, followed by cell lysis and luciferase activity assays. Promoter constructs were transactivated by the wild-type cotransfected RORα1, -α4, or -γ1 expression plasmids significantly compared with empty vector (***, p < 0.001) but not the RORα4 DNA binding mutant and only slightly by the RORγ mutant lacking the AF2 domain. In contrast, transactivation was abolished when the -2045RORE was mutated. Values represent the means of three independent transfections ± S.E.
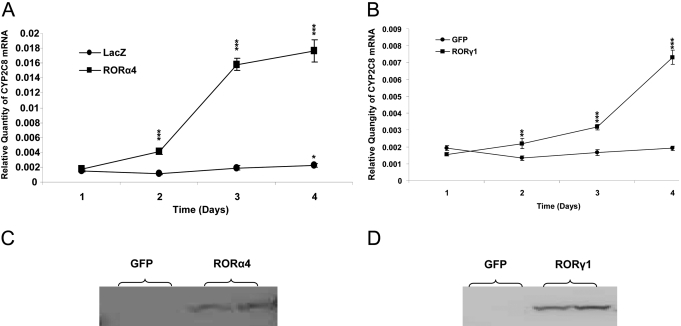
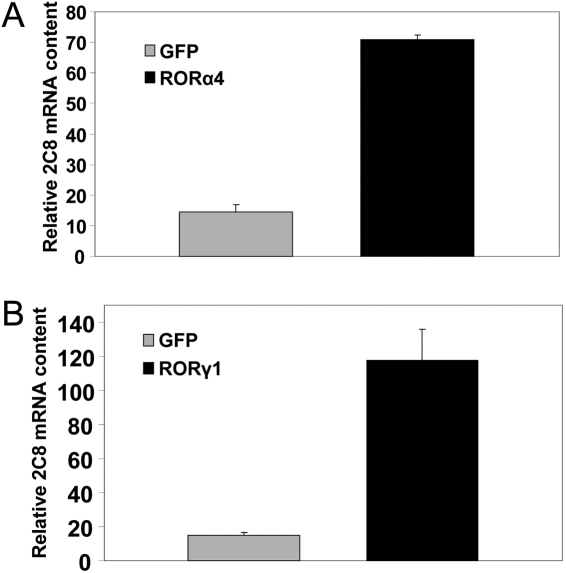
Endogenous CYP2C8 Was Elevated by Exogenous RORs in HepG2 Cells and in Human Primary Hepatocytes. We introduced exogenous murine ROR proteins into HepG2 cells to investigate their effects on the expression of endogenous CYP2C8. We prepared adenoviral constructs to overexpress wild-type mROR protein in HepG2 cells. CYP2C8 expression was measured for 4 days after infection. As confirmed in Western blots (Fig. 5, C and D), both the wild-type RORα4 and -γ1 proteins were overproduced in HepG2 cells. Compared with the LacZ control, adenoviral overexpressed RORα4 significantly increased the level of endogenous CYP2C8 mRNA (p < 0.001) in a time-dependent manner. By the 4th day after infection, a 9-fold increase in the CYP2C8 mRNA level was observed compared with cells infected with LacZ (p < 0.001) (Fig. 5A). Overexpression of RORγ1 also increased endogenous CYP2C8 mRNA, reaching a 4-fold higher level by day 4 compared with the control adenovirus expressing GFP (p < 0.001). No change in CYP2C8 transcripts was observed in cells infected with the GFP control (Fig. 5B).
Fig. 5.
Overexpression of murine RORα4 and γ1 dramatically increases endogenous CYP2C8 mRNA in HepG2 cells. Adenoviruses containing FLAG-mRORα4 and the control LacZ (A) or FLAG-mRORγ1 and the control GFP/β-Gal (B) were utilized to infect HepG2 cells in triplicate for 1 to 4 days with a dose of 1000 particles/cell. Cells were harvested and used to isolate total RNA. cDNAs were synthesized with MMLV, and then real-time quantitative PCR analyses were performed to determine the expression of CYP2C8 and TBP. Endogenous CYP2C8 mRNA gradually increased after infection of HepG2 cells with adenoviruses containing ROR over those infected with LacZ significantly (**, p < 0.01; ***, p < 0.001). Data represent means ± S.E. (n = 9). All data are normalized to TBP levels. The same ROR viruses and the GFP control were used to infect HepG2 cells in six-well plate in duplicates. After 2 days, the whole-cell lysates were prepared, and the Western blot was performed to detect the expression of exogenous FLAG-tagged ROR protein (C and D).
Next, we examined whether exogenous RORs could elevate CYP2C8 expression in primary human hepatocytes. Adenoviruses expressing wild-type murine RORα4 and RORγ1 were used to infect primary human hepatocytes for 60 h. The relative amount of endogenous hRORα mRNA (relative to TBP) was 5.2 ± 0.1 in GFP-infected cells. After transfection, the level of mRORα expression was 200-fold higher (1209 ± 131) than that of endogenous hRORα. The amount of endogenous hRORγ in GFP-infected cells was 1.8 ± 0.2, whereas after transfection with mRORγ, RORγ levels were ∼24 times higher (44 ± 5). It should be noted that amino acid identity between human and murine RORs is ∼97% for RORα proteins and 89% for RORγ proteins, suggesting their biological effects should be very similar. Endogenous CYP2C8 mRNA was significantly increased, 5-fold by exogenous RORα4 and 7-fold by exogenous RORγ1 (p < 0.001) (as shown in Fig. 6, A and B). The effects of exogenous RORα and RORγ on CYP2C8 expression in HepG2 cells and primary hepatocytes suggest that RORs can activate the endogenous CYP2C8 gene in human liver.
Fig. 6.
Overexpression of murine ROR dramatically elevated endogenous CYP2C8 mRNA in human primary hepatocytes. Human primary hepatocytes were infected with adenoviruses for FLAG-mRORα4 (A) and FLAG-mRORγ1 (B) and the control GFP/β-Gal at a dose of 1000 particles/cell. Total RNA was isolated after 60 h, and real-time quantitative PCR analyses were performed to determine the expression of CYP2C8 relative to TBP. Both ROR viruses significantly increased endogenous CYP2C8 mRNA over the GFP adenoviral control (p < 0.001). Data represent means ± S.E. (n = 9). All data are normalized to TBP levels.
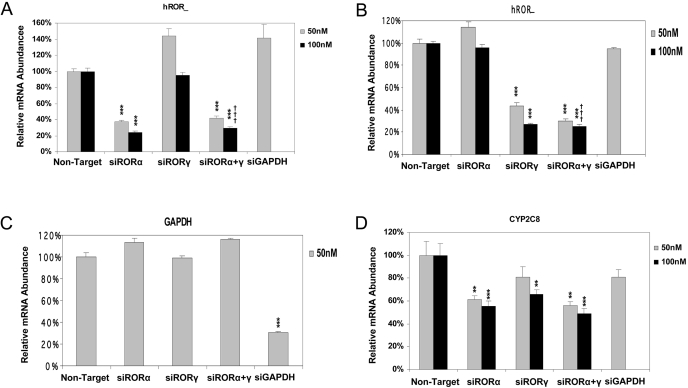
Endogenous CYP2C8 Was Reduced by the Decrease in Endogenous RORs in HepG2 Cells. To gain more insight into the involvement of RORs in the transcriptional regulation of the CYP2C8 gene in vivo, we used siRNA technology to knock down endogenous ROR expression and examined its effect on endogenous CYP2C8 mRNA expression. siRNA oligos against human RORα and RORγ were transfected into HepG2 cells individually or in combination at low and high doses. As shown in Fig. 7, compared with transfection with the nontarget oligo, only the siRNAs that are specific for their target genes (RORα, RORγ, and GAPDH) could significantly reduce the mRNA levels of their targets; the siRNA for RORα reduced endogenous RORα mRNA specifically to 30% at the high dose 100 nM either used individually or in combination with siRNA for RORγ (p < 0.001) (Fig. 7A). Endogenous RORγ mRNA was significantly reduced to 30% exclusively by its siRNA at the high dose (p < 0.001) (Fig. 7B). RORα and RORγ were decreased significantly more at the high dose of 100 nM siRNA than the low dose of 50 nM (p < 0.001). In accordance, CYP2C8 mRNA was reduced to 60% with the transfection of siRNA for RORα alone at both doses (p < 0.01 with 50 nM and p < 0.001 with 100 nM) and to 65% with the transfection of siRNA for RORγ at the high dose of 100 nM (p < 0.01). However, it should be noted that the level of RORα mRNA is approximately 3-fold higher than that of RORγ in HepG2 control cells, which is consistent with the slightly greater effect of knockdown of RORα on CYP2C8 mRNA concentrations. When the two siRNAs were given in combination, CYP2C8 mRNA concentrations were reduced to 57% at 50 nM (p < 0.01) or 50% at 100 nM (p < 0.001). siRNA for GAPDH did not significantly affect the expression of CYP2C8 transcripts when transfected at 50 nM (Fig. 7D).
Fig. 7.
Reduction of endogenous human RORs by using siRNA oligos significantly decreased endogenous CYP2C8 mRNA in HepG2 cells. Individual or combinational siRNA oligos were transfected into HepG2 cells at two doses of 50 and 100 nM, respectively. Total RNAs were prepared from those transfected cells and subjected to RT-PCR to measure the mRNA levels for hRORα (A), hRORγ (B), GAPDH (C), and CYP2C8 (D). The siRNA oligos specifically decreased the mRNA level of their corresponding target genes and CYP2C8, comparing with the nontarget control significantly (**, p < 0.01; ***, p < 0.001), whereas the higher dose of siRNA oligos yielded a larger decrease compared with the lower dose (†††, p < 0.001). Data represent average ± S.E. (n = 9). All data are normalized to TBP levels.
Discussion
In the present study, we show that the 3 kb of the human proximal CYP2C8 promoter is specifically activated by RORα1, RORα4, and RORγ1 in HepG2 cells, the forms of ROR known to be expressed in human liver. In contrast, neither the CYP2C9 nor CYP2C19 promoters are activated by RORs, which was confirmed with both 3- and 12-kb promoter constructs. Two putative ROREs were identified and shown to interact specifically with RORα1, -α4, and -γ1 proteins in gel shift assays; however, the proximal site at -2045 bp showed a much stronger affinity for all three isoforms than the more distal site. Mutation of the ROREs showed that only the proximal site plays a role in activation of the CYP2C8 promoter by RORs. In addition, endogenous CYP2C8 mRNA expression was up-regulated by exogenous RORα and -γ proteins in both HepG2 cells and primary hepatocytes. siRNA studies were able to achieve 70% knockdown of RORα and RORγ isoforms in HepG2 cells in the present study. Double knockdown of RORα and -γ isoforms produced a 50% decrease in CYP2C8 mRNA. These knockdown studies might underestimate the role of RORs in liver somewhat because of the fact the knockdown of RORs receptors was not complete. The present study indicates that the ROR nuclear receptor family plays a role in regulating the expression of the human P450 gene CYP2C8 in liver and perhaps other tissues. In addition, constitutive levels of CYP2C8 are probably regulated to some extent by HNF4α in liver (Ferguson et al., 2005).
The CYP2C8 enzyme oxidizes a variety of structurally unrelated compounds, including steroids, fatty acids, and xenobiotics. It is important in the metabolism of arachidonic acid to physiologically active compounds, such as epoxyeicosatrienoic acids (EETs) (Zeldin, 2001). CYP2C8 is the principal enzyme responsible for the metabolism of clinically important drugs, such as the anticancer drug paclitaxel, the antimalarial drug amodiaquine, the antidiabetic drugs troglitazone and rosiglitazone, the antiarrhythmic drug amiodarone, and the calcium channel blocker verapamil (Totah and Rettie, 2005). The discovery that CYP2C8 is transactivated by RORα and -γ in hepatocytes suggests the possibility that the clearance of these drugs might be modulated to some extent by ligands of RORs that modulate ROR activity. Recent X-ray crystal structure studies have demonstrated a series of natural compounds that can reversibly bind to RORα and act as agonists increasing its transactivational activity (Kallen et al., 2002, 2004). These include cholesterol and its structural derivatives 7-dehydrocholesterol and cholesterol sulfates. For example, depletion of cholesterol by the drug lovastatin in the osteosarcoma cells U-20S in vitro has been proposed to modulate RORα transcriptional activity (Kallen et al., 2002). In addition, certain natural compounds, including several retinoids such as all-trans-retinoic acid and the synthetic retinoid ALRT1550 (Ligand Pharmaceuticals, San Diego, CA), can bind RORβ and RORγ and act as partial antagonists, inhibiting their transactivation activity (Stehlin-Gaon et al., 2003).
Recent studies have implicated RORs in the control of circadian rhythm, both in the central nervous system (RORα and -β) (Jetten and Joo, 2006) and in peripheral tissues such as liver (RORα and -γ) (Jetten and Joo, 2006). In mammals, many physiological and behavioral processes exhibit daily oscillations, including many hepatic enzyme activities, including those involved in energy metabolism, and a clear link between the control of circadian rhythm and metabolism has been established (Albrecht, 2006; Hastings et al., 2007). The clock oscillator consists of interlocked positive and negative transcriptional/posttranscriptional feedback loops between the clock genes Clock/Bmal1 and Per/Crys. By binding to the RORE within target gene promoters, RORs, along with the negative competitors Rev-Erbα and -β, control the expression of Bmal1 (Jetten and Joo, 2006). A number of rodent P450 enzymes display a daily fluctuation in their mRNA expression or catalytic activity, such as Cyp1a1 and 1b1, Cyp2a, Cyp2c, Cyp2e1, Cyp4a, Cyp7a1 and 7b1, and Cyp8b1 (Kang et al., 2007; Ohdo, 2007). RORα has been suggested to be involved in the oscillatory regulation of some of these P450 enzymes (Wada et al., 2008). In humans, the effect of circadian rhythm on drug metabolism has not been studied extensively. However, studies from Ohdo and colleagues have suggested that the effectiveness of drugs depends on the time of day at which they are administered (Kang et al., 2007; Ohdo, 2007). Moreover, they showed that activity of CYP3A4 exhibits an ∼2.8-fold diurnal variation in humans based on 6-hydroxycortisol/cortisol ratios, which are a noninvasive index of CYP3A4 activity (Ohdo, 2007). It would not be surprising to find an oscillatory regulation in other human P450 enzymes such as CYP2C8 because the expression of a number of nuclear receptors, including RORs and HNF4α, CAR, peroxisome proliferator-activated receptor, estrogen-related receptor, and small heterodimer partner, display circadian rhythm (Yang et al., 2006).
CYP2C8 is the one CYP2C member that has the broadest tissue expression. It has been found to be expressed primarily in liver but also exists in many other tissues such as brain, heart, endothelial cells, intestine, and kidney (Klose et al., 1999). Both RORα and RORγ are widely expressed in most of these tissues (Jetten and Joo, 2006). This overlapping distribution is consistent with the possibility that RORs may also regulate CYP2C8 in some of these extrahepatic tissues. In brain, RORα is expressed in many regions, including the thalamus and cerebellum, where it plays an important role in circadian rhythm. RORβ is also found in brain. CYP2C8 mRNA has been detected at relatively high levels in many regions in human brain, including cerebellum, although unlike ROR, the distribution of CYP2C8 has not been studied in different cell types in the brain using immunohistochemical approaches (McFayden et al., 1998; Klose et al., 1999). RORα is known to be expressed in small intestinal epithelium and in the human colon cell line Caco-2 (Jetten and Joo, 2006). The present study demonstrates that the CYP2C8 promoter can be activated by RORα1 in Caco-2 cells, indicating a possible regulatory role of RORs in transcription of CYP2C8 in colon and intestine. mRNA of RORα also has been detected in endothelial cells (Besnard et al., 2002), where CYP2C8 oxidizes arachidonic acid to produce 11,12- and 14,15-EETs that have vasodilatory roles and anti-inflammatory roles (Wray and Bishop-Bailey, 2008). If CYP2C8 expression is up-regulated by RORα1 in endothelial cells, this might increase EET formation, thus enhancing dilation and producing anti-inflammatory responses.
In summary, in the present study, we provide evidence demonstrating that RORα1, -α4, and -γ1 positively regulate CYP2C8 gene expression in human hepatocytes through an RORE in the CYP2C8 proximal promoter region. Because CYP2C8 catalyzes the metabolism of a number of clinically important drugs, and RORs function as ligand-dependent transcription factors, ROR (ant)agonists may be able to control the expression of CYP2C8 and, therefore, the metabolism of these drugs. In addition, because RORs play a role in the regulation of circadian rhythm, they might play a role in the possible circadian regulation of CYP2C8.
Acknowledgments
We thank Dr. Ritu Rana and Dr. Sailesh Surapureddi (Laboratory of Pharmacology, National Institute of Environmental Health Sciences) for assistance in preparation of the adenovirus that was used in our studies and for helpful discussions; other members of our team, including Joyce Blaisdell (National Institute of Environmental Health Sciences) for Ad-293 cell culture and other technical support; Gary Burke (Summers of Discovery Program, National Institute of Environmental Health Sciences) for preparing some CYP2C8 constructs; Dr. Stephen Ferguson (CellzDirect) for a gift of some of the human primary hepatocytes used in the study; and Drs. Masa Negishi and Hong Soon Kang (National Institute of Environmental Health Sciences) for comments regarding the manuscript.
This work was supported by the Intramural Research Program of the National Institutes of Health National Institute of Environmental Health Sciences.
doi:10.1124/jpet.108.148916.
ABBREVIATIONS: P450, cytochrome P450; CAR, constitutive androstane receptor; PXR, pregnane X receptor; HNF, hepatic nuclear factor; ROR, retinoid-related orphan nuclear receptor; RORE, ROR-responsive element; PCR, polymerase chain reaction; RT, reverse transcriptase; GFP, green fluorescent protein; β-Gal, β-galactosidase; TBP, TATA-box binding protein; GAPDH, glyceraldehyde-3-phosphate dehydrogenase; siRNA, small interference RNA; m-pcp-2, murine Purkinje cell protein-2; bp, base pair; EET, epoxyeicosatrienoic acid.
References
- Albrecht U (2006) Orchestration of gene expression and physiology by the circadian clock. J Physiol Paris 100 243-251. [DOI] [PubMed] [Google Scholar]
- Besnard S, Heymes C, Merval R, Rodriguez M, Galizzi JP, Boutin JA, Mariani J, and Tedgui A (2002) Expression and regulation of the nuclear receptor RORalpha in human vascular cells. FEBS Lett 511 36-40. [DOI] [PubMed] [Google Scholar]
- Bort R, Gómez-Lechón MJ, Castell JV, and Jover R (2004) Role of hepatocyte nuclear factor 3 gamma in the expression of human CYP2C genes. Arch Biochem Biophys 426 63-72. [DOI] [PubMed] [Google Scholar]
- Chauvet C, Bois-Joyeux B, and Danan JL (2002) Retinoic acid receptor-related orphan receptor (ROR) alpha4 is the predominant isoform of the nuclear receptor RORalpha in the liver and is up-regulated by hypoxia in HepG2 human hepatoma cells. Biochem J 364 449-456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chauvet C, Bois-Joyeux B, Fontaine C, Gervois P, Bernard MA, Staels B, and Danan JL (2005) The gene encoding fibrinogen-beta is a target for retinoic acid receptor-related orphan receptor alpha. Mol Endocrinol 19 2517-2526. [DOI] [PubMed] [Google Scholar]
- Chen Y, Ferguson SS, Negishi M, and Goldstein JA (2003) Identification of constitutive androstane receptor and glucocorticoid receptor binding sites in the CYP2C19 promoter. Mol Pharmacol 64 316-324. [DOI] [PubMed] [Google Scholar]
- Chen Y, Ferguson SS, Negishi M, and Goldstein JA (2004) Induction of human CYP2C9 by rifampicin, hyperforin, and phenobarbital is mediated by the pregnane X receptor. J Pharmacol Exp Ther 308 495-501. [DOI] [PubMed] [Google Scholar]
- Chen Y, Kissling G, Negishi M, and Goldstein JA (2005) The nuclear receptors constitutive androstane receptor and pregnane X receptor cross-talk with hepatic nuclear factor 4alpha to synergistically activate the human CYP2C9 promoter. J Pharmacol Exp Ther 314 1125-1133. [DOI] [PubMed] [Google Scholar]
- Ferguson SS, Chen Y, LeCluyse EL, Negishi M, and Goldstein JA (2005) Human CYP2C8 is transcriptionally regulated by the nuclear receptors constitutive androstane receptor, pregnane X receptor, glucocorticoid receptor, and hepatic nuclear factor 4alpha. Mol Pharmacol 68 747-757. [DOI] [PubMed] [Google Scholar]
- Genoux A, Dehondt H, Helleboid-Chapman A, Duhem C, Hum DW, Martin G, Pennacchio LA, Staels B, Fruchart-Najib J, and Fruchart JC (2005) Transcriptional regulation of apolipoprotein A5 gene expression by the nuclear receptor RORalpha. Arterioscler Thromb Vasc Biol 25 1186-1192. [DOI] [PubMed] [Google Scholar]
- Gerbal-Chaloin S, Daujat M, Pascussi JM, Pichard-Garcia L, Vilarem MJ, and Maurel P (2002) Transcriptional regulation of CYP2C9 gene: role of glucocorticoid receptor and constitutive androstane receptor. J Biol Chem 277 209-217. [DOI] [PubMed] [Google Scholar]
- Goldstein JA (2001) Clinical relevance of genetic polymorphisms in the human CYP2C subfamily. Br J Clin Pharmacol 52 349-355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Handschin C and Meyer UA (2003) Induction of drug metabolism: the role of nuclear receptors. Pharmacol Rev 55 649-673. [DOI] [PubMed] [Google Scholar]
- Hastings M, O'Neill JS, and Maywood ES (2007) Circadian clocks: regulators of endocrine and metabolic rhythms. J Endocrinol 195 187-198. [DOI] [PubMed] [Google Scholar]
- Jetten AM and Joo JH (2006) Retinoid-related Orphan Receptors (RORs): Roles in Cellular Differentiation and Development. Adv Dev Biol 16 313-355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jover R, Bort R, Gómez-Lechón MJ, and Castell JV (1998) Re-expression of C/EBP alpha induces CYP2B6, CYP2C9 and CYP2D6 genes in HepG2 cells. FEBS Lett 431 227-230. [DOI] [PubMed] [Google Scholar]
- Jover R, Bort R, Gómez-Lechón MJ, and Castell JV (2001) Cytochrome P450 regulation by hepatocyte nuclear factor 4 in human hepatocytes: a study using adenovirus-mediated antisense targeting. Hepatology 33 668-675. [DOI] [PubMed] [Google Scholar]
- Kallen J, Schlaeppi JM, Bitsch F, Delhon I, and Fournier B (2004) Crystal structure of the human RORalpha Ligand binding domain in complex with cholesterol sulfate at 2.2 A. J Biol Chem 279 14033-14038. [DOI] [PubMed] [Google Scholar]
- Kallen JA, Schlaeppi JM, Bitsch F, Geisse S, Geiser M, Delhon I, and Fournier B (2002) X-ray structure of the hRORalpha LBD at 1.63 A: structural and functional data that cholesterol or a cholesterol derivative is the natural ligand of RORalpha. Structure 10 1697-1707. [DOI] [PubMed] [Google Scholar]
- Kang HS, Angers M, Beak JY, Wu X, Gimble JM, Wada T, Xie W, Collins JB, Grissom SF, and Jetten AM (2007) Gene expression profiling reveals a regulatory role for ROR alpha and ROR gamma in phase I and phase II metabolism. Physiol Genomics 31 281-294. [DOI] [PubMed] [Google Scholar]
- Klose TS, Blaisdell JA, and Goldstein JA (1999) Gene structure of CYP2C8 and extrahepatic distribution of the human CYP2Cs. J Biochem Mol Toxicol 13 289-295. [DOI] [PubMed] [Google Scholar]
- Komoroski BJ, Zhang S, Cai H, Hutzler JM, Frye R, Tracy TS, Strom SC, Lehmann T, Ang CY, Cui YY, et al. (2004) Induction and inhibition of cytochromes P450 by the St. John's wort constituent hyperforin in human hepatocyte cultures. Drug Metab Dispos 32 512-518. [DOI] [PubMed] [Google Scholar]
- Madan A, Graham RA, Carroll KM, Mudra DR, Burton LA, Krueger LA, Downey AD, Czerwinski M, Forster J, Ribadeneira MD, et al. (2003) Effects of prototypical microsomal enzyme inducers on cytochrome P450 expression in cultured human hepatocytes. Drug Metab Dispos 31 421-431. [DOI] [PubMed] [Google Scholar]
- McFayden MC, Melvin WT, and Murray GI (1998) Regional distribution of individual forms of cytochrome P450 mRNA in normal adult human brain. Biochem Pharmacol 55 825-830. [DOI] [PubMed] [Google Scholar]
- Medvedev A, Yan ZH, Hirose T, Giguère V, and Jetten AM (1996) Cloning of a cDNA encoding the murine orphan receptor RZR/ROR gamma and characterization of its response element. Gene 181 199-206. [DOI] [PubMed] [Google Scholar]
- Ohdo S (2007) Chronopharmacology focused on biological clock. Drug Metab Pharmacokinet 22 3-14. [DOI] [PubMed] [Google Scholar]
- Raucy JL, Mueller L, Duan K, Allen SW, Strom S, and Lasker JM (2002) Expression and induction of CYP2C P450 enzymes in primary cultures of human hepatocytes. J Pharmacol Exp Ther 302 475-482. [DOI] [PubMed] [Google Scholar]
- Stehlin-Gaon C, Willmann D, Zeyer D, Sanglier S, Van Dorsselaer A, Renaud JP, Moras D, and Schüle R (2003) All-trans retinoic acid is a ligand for the orphan nuclear receptor ROR beta. Nat Struct Biol 10 820-825. [DOI] [PubMed] [Google Scholar]
- Totah RA and Rettie AE (2005) Cytochrome P450 2C8: substrates, inhibitors, pharmacogenetics, and clinical relevance. Clin Pharmacol Ther 77 341-352. [DOI] [PubMed] [Google Scholar]
- Wada T, Kang HS, Angers M, Gong H, Bhatia S, Khadem S, Ren S, Ellis E, Strom SC, Jetten AM, et al. (2008) Identification of oxysterol 7alpha-hydroxylase (Cyp7b1) as a novel retinoid-related orphan receptor alpha (RORalpha) (NR1F1) target gene and a functional cross-talk between RORalpha and liver X receptor (NR1H3). Mol Pharmacol 73 891-899. [DOI] [PubMed] [Google Scholar]
- Wray J and Bishop-Bailey D (2008) Epoxygenases and peroxisome proliferator-activated receptors in mammalian vascular biology. Exp Physiol 93 148-154. [DOI] [PubMed] [Google Scholar]
- Yang X, Downes M, Yu RT, Bookout AL, He W, Straume M, Mangelsdorf DJ, and Evans RM (2006) Nuclear receptor expression links the circadian clock to metabolism. Cell 126 801-810. [DOI] [PubMed] [Google Scholar]
- Zeldin DC (2001) Epoxygenase pathways of arachidonic acid metabolism. J Biol Chem 276 36059-36062. [DOI] [PubMed] [Google Scholar]