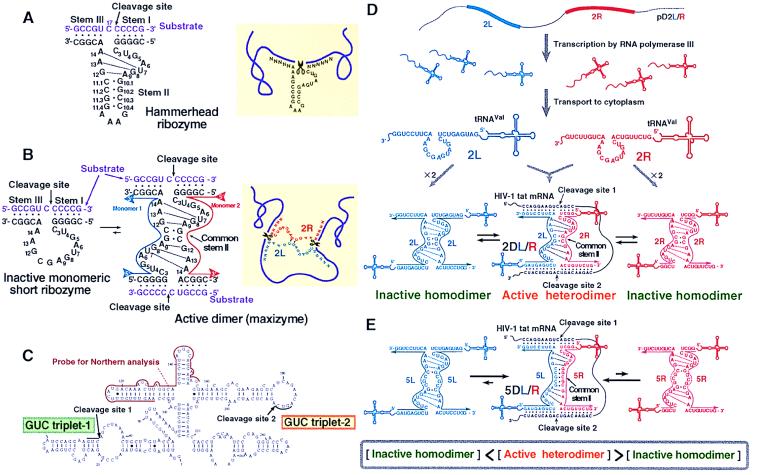
Figure 1.
Secondary structures of (A) the wild-type hammerhead ribozyme and (B) the minizyme (Left) that is capable of forming a homodimer [maxizyme (Center)]. All ribozymes were γ-shaped in the crystals, with stems I and II forming the arm of the γ and stem III forming the base and with stem I and stem II being adjacent to each other and stems II and III being stacked colinearly to form a pseudo-A-form helix (2). As shown in A by dotted lines, the nucleotides within the catalytic loop form two reversed-Hoogsteen G-A base pairs between G8-A13 and A9-G12 and a non-Watson–Crick A14-U7 base pair that consists of one hydrogen bond. These additional base pairs strengthen the dimeric form of the maxizyme (B). The heterodimeric maxizymes can cleave an mRNA at two sites simultaneously (B Right). Secondary structure of tat mRNA is shown in C. (D) The active heterodimeric and inactive homodimeric forms of 2-bp dimeric maxizymes under the control of a human tRNAVal-promoter. A large fraction of the population of dimers is expected to be in the inactive homodimeric forms. (E) Active heterodimeric and inactive homodimeric forms of the 5-bp dimeric maxizymes. The formation of active forms is favored because perfect base pairing occurs only in the case of active complexes.