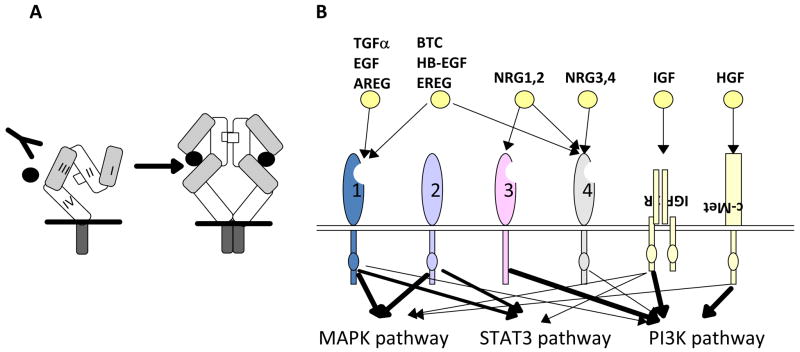
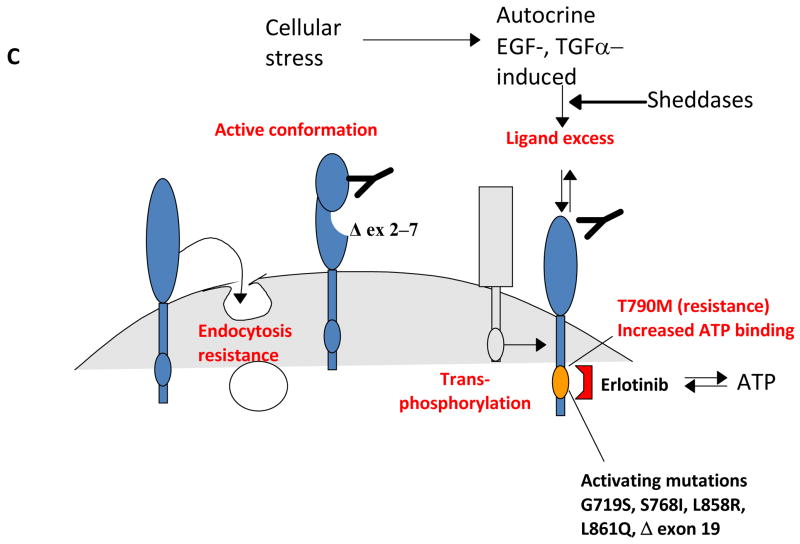
Figure 1. EGFR/(v-erb-b) oncogene homolog 1 (ErbB1) and the ErbB protein family.
A, EGFR structure. Defined protein domains are numbered I‐IV [113]. Binding of ligand (black circles) to domains I and III of EGFR induces unfolding from a tethered to an ‘activated’ conformation. The resulting exposure of dimerization domain II allows it to pair with another EGFR molecule, or heterodimerize with another ErbB family member. Intracellular juxtaposition of the EGFR kinase domains induces trans-phosphorylation of essential tyrosine residues, activating downstream signaling. Cetuximab targets domain III, preventing ligand binding and subsequent activation steps. B. The ErbB receptor family, collaborating receptors, and receptor ligands [10]. EGFR homodimerizes, and heterodimerizes with all other ErbB family members [219]. All family members except ErbB3 contain a kinase domain (small ovals); ErbB3 is active in downstream signaling because of transphosphorylation. The profile of ligand binding to specific ErbB family members is indicated. AREG: amphiregulin; BTC: betacellulin; EREG: epiregulin; HB-EGF: heparin-binding EGF-like growth factor; HGF: hepatocyte growth factor; NRG: neuregulin; PI3K: phosphinositol 3 kinase; STAT :Signal transducer and activator of transcription. Signals emanating from c-Met, IGF-1R and ErbB3 modulate similar downstream outputs as ErbB family members: see text for details. C. Mechanisms of resistance to anti-EGFR agents. Deletion of EGFR exons 2-7 leads to constitutively active conformation of the EGFR [101,104]. EGFR kinase domain mutations, such as the common T790M mutation [86], may result in increased ATP binding and resistance to erlotinib. Receptor non-intrinsic mechanisms of resistance to anti-EGFR agents include ligand excess [115], resistance to antibody-induced endocytosis [16], or transphosphorylation [129,130].