Figure 3.
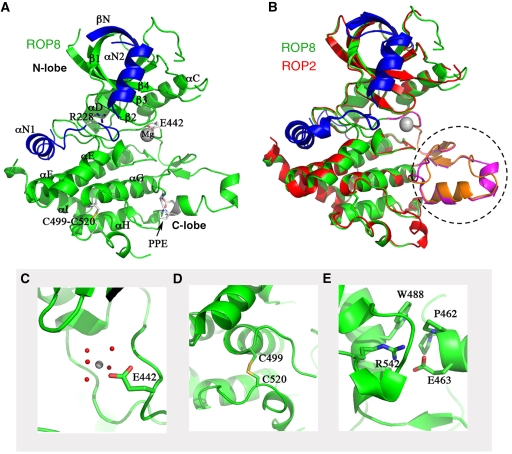
Models of ROP2 and ROP8 structures. (A) Ribbon model of ROP8 structure shows a typical kinase fold consisting of N- and C-lobes. N-terminal extension provides two additional alpha helices (αN1, αN2) and an extra beta sheet (βN) capping the N-lobe (blue). Major helices, including αC, are labelled, as are the magnesium ion, the inhibitory arginine (R228), the intrachain disulphide bond (C499–C520), Glu442 (E442) and the PPE motif at the end of the activation loop. (B) Superposition of ROP2 (red) on ROP8 (green) shows a tight alignment, including both the N- and C-lobes and the N-terminal regulatory domain. Major region of difference is in the activation loop and substrate-binding region (dotted circle, magenta=ROP8, orange=ROP2). (C) Enlarged view of Glu442 in ROP8 that coordinates the Mg2+ ion. (D) Intrachain disulphide bond in ROP8 stabilizes some of the C-lobe helices. (E) Enlarged view of the PPE motif stabilizing the substrate-binding region.