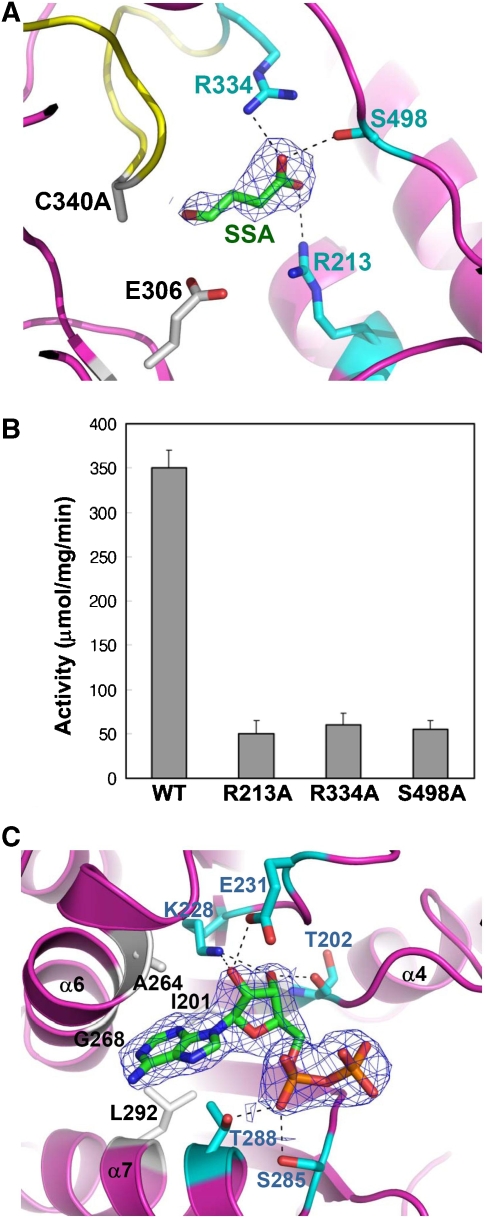
Figure 4.
Substrate- and cofactor-binding properties of human SSADH. (A) Diagram showing the SSA-binding site. To prevent the enzyme reaction and keep the enzyme in ‘open' formation, the C340A mutant was used for the complex crystallization. The C340A mutant is shown as a ribbon model in magenta colour, and its ‘dynamic catalytic loop' is shown in yellow. SSA is shown as a stick model in green colour, and the 2Fo−Fc electron density (blue mesh) of SSA is contoured at 0.8σ. Residues, whose side chains form hydrogen bonds to the carboxyl group of SSA, are shown as stick models in cyan colour, and the hydrogen bond interactions are indicated by black dotted lines. The catalytic residues, E306 and C340 (substituted with alanine in the present structure), are shown as stick models in grey colour. (B) SSADH activity assays. Each of three residues (R213, R334 and S498) involved in the SSA binding was substituted with alanine by site-directed mutagenesis, and SSADH activity assays were performed by measuring the reduction of NAD+ to NADH. (C) SSADH–NAD complex structure. The 2Fo−Fc electron density (blue mesh) of ADP moiety of NAD+ molecule is contoured at 1.0σ. The residues involved in hydrogen bond formations with ADP moiety and in forming hydrophobic cavity are shown as a stick model in cyan and grey colours, respectively. The hydrogen bond interactions are indicated by black dotted lines.