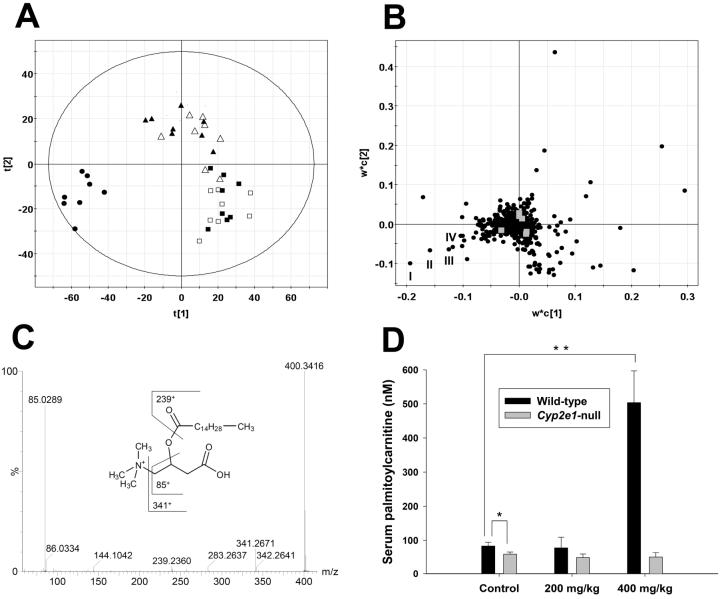
Figure 1.
Identification of acylcarnitines as serum biomarkers of APAP toxicity through metabolomic analysis of 24-h serum samples from wild-type and Cyp2e1-null mice treated with 200 and 400 mg/kg APAP. A, Scores scatter plot of an PLS-DA model on the APAP-elicited dose-dependent influence on the serum metabolome. Details of data processing and model construction were described in the Experimental procedures. A two-component PLS-DA model was constructed to characterize the relationship among six mouse groups (n=7 or 8 mice/group), including wild-type mice (control: ■; 200 mg/kg APAP: ▲; 400 mg/kg APAP: •) and Cyp2e1-null mice (control: □; 200 mg/kg APAP: Δ; 400 mg/kg APAP: ○). The t[1] and t[2] values represent the scores of each sample in principal component 1 and 2, respectively. Fitness (R2) and prediction power (Q2) of this PLS-DA model are 0.486 and 0.429, respectively. The model was validated through the recalculation of R2 and Q2 values after the permutation of sample identities. B, Loadings scatter plot representing the correlation between individual serum ion (w*) and each sample group (c) in the 1st and 2nd components of the PLS-DA model. Data points representing palmitoylcarnitine (I), myristoylcarnitine (II), oleoylcarnitine (III) and palmitoleoylcarnitine (IV) were labeled in the plot. C, MS2 fragmentation of palmitoylcarnitine (I). Major fragment ions were interpreted in the inlaid structural diagrams. D, Influence of APAP treatment on serum palmitoylcarnitine level (mean ± SD) in wild-type and Cyp2e1-null mice (n=4; *, p < 0.05 and **, p < 0.01). Palmitoylcarnitine level in serum (mean ± SD) was measured using the multiple reactions monitoring mode in LC-MS. [2H3]palmitoylcarnitine was used as internal standard.