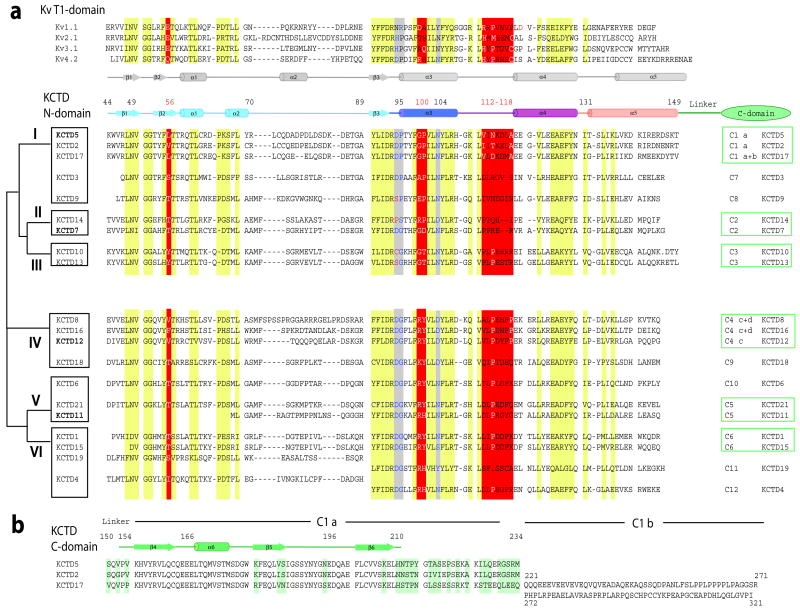
Figure 3. Sequence and secondary structure alignments.
KCTD BTB domains share 33% to 43% sequence homology with T1 domains of Kv channels 1,2 with most differences in the middle and C-terminus of the fold. Zinc binding motifs in T1 domains of Kv2, Kv3 and Kv4 channels are not found in KCTDs.
a. Alignment of T1-domains of select Kv channels and KCTD N-terminal BTB domains based on X-ray structures: KCTD5 (this work), Kv1.1 (PDBid 1T1D), Kv3.1 (PDBid 3KVT), Kv4.2 (PDBid 1NN7). Secondary structure elements are those determined for Kv4.2 and KCTD5. Conserved patches are highlighted in yellow. Key differences are highlighted in red. The alignment of KCTD proteins was by ClustalW. KCTD20 is omitted. The schematic on left is a KCTD group classification. Residues before the BTB domain are omitted. The KCTD5 N-terminal BTB fold is 106 residues long (residues 44–149). The consensus sequence (including insertions) is 110 amino acids with 26 residues in the diverse α2-β3 loop (corresponds to KCTD5 residues 70–89) and 14 residues in α5 (KCTD5 residues 132–149). Sequences of the α2-β3 loop and α5-helix display homology only between proteins of the same group.
The C-termini of 14 KCTDs show significant identity with at least one other variant while seven are distinct. The % similarity of groups C1, C2, C3, C4, C5, and C6 are 88, 50, 93, 69, 48, and 83, respectively.
b. Alignment of KCTD5, KCTD2 and KCTD17 C-termini; differences are highlighted in green. Residues 212–234 are disordered in the KCTD5 crystal structure.