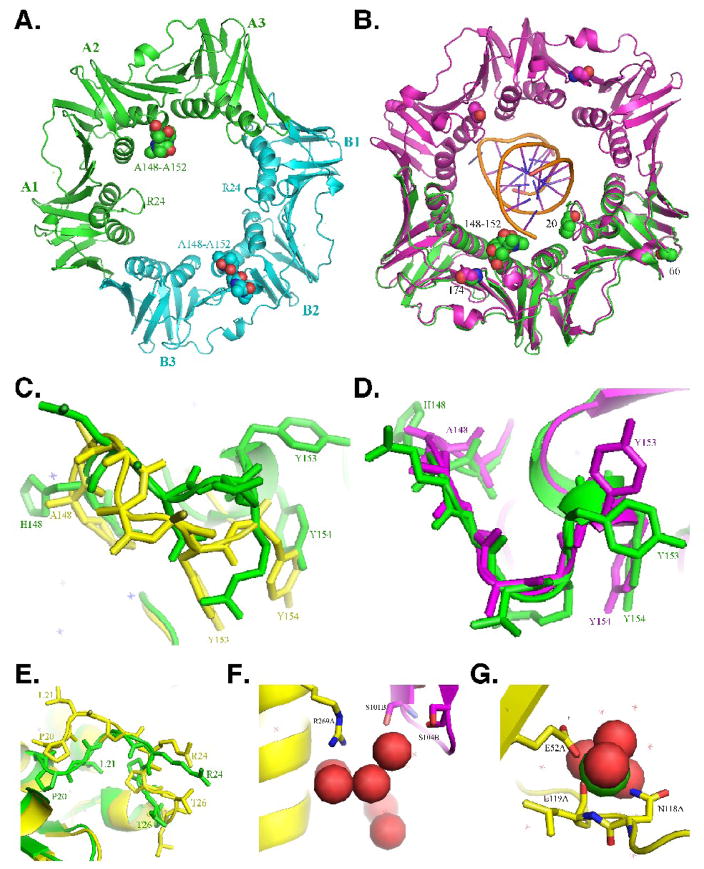
FIGURE 1. X-ray crystal structure of the mutant β148–152 clamp protein.
(A) Face view (C-side) of the structure of the mutant β148–152 clamp protein depicted in ribbon form. The three subdomains comprising each protomer (A1–3, and B1–3) are indicated, as are the positions of the loops containing the 148-HQDVR-152 poly-Ala substitutions (150; depicted as space filled atoms), and residues L21–L27 (24; depicted in ribbon form). Protomer A is in green, and protomer B is in blue. (B) Face view (C-side) of the wild type β clamp protein (purple) encircling primed DNA, the backbone of which is shown in gold (PDB: 3BEP)11. Protomer A of β148–152 from panel A (green) is superimposed onto protomer A of the wild type clamp (purple). Positions of the 148-HQDVR-152 poly-Ala substitutions, G66 and G174, which are substituted with E and G, respectively, in β159, as well as P20, which is substituted with L in dnaN783, and acts to suppress the temperature sensitive growth phenotype of the dnaN159 allele,25 are indicated. Enlarged views of the loop containing residues 148-HQDVR-152 in protomer A (C) or B (D) are shown. Wild type β (PDB: 1OK7)20 is depicted in green, while β148–152 is in yellow (C) or purple (D). (E) Enlarged view of the loop containing residues L21–L27. Protomer A of wild type β (PDB: 1OK7) is depicted in yellow, while protomer A of β148–152 is in green. (F) Enlarged view of the water network near the β148–152 dimer interface. Positions R269 of protomer A (yellow), and S101 and S104 of protomer B (purple) are indicated. Water is depicted as space filled atoms. (G) Enlarged view of the interaction of protomer A of β148–152 with calcium present in the crystallization buffer. Positions of resides E52, N118, and L119 are indicated. Calcium is depicted as space filled atoms.