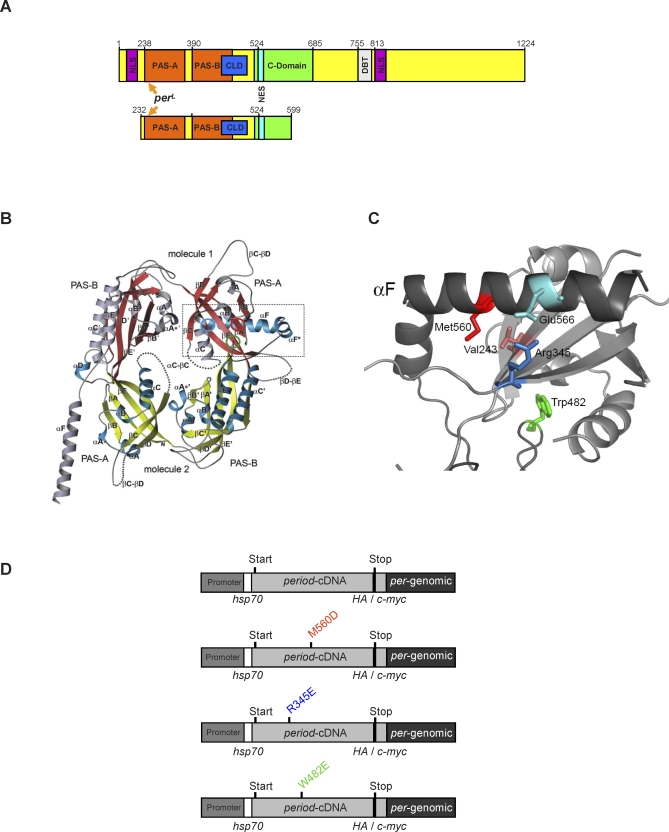
Figure 1. Structure of the PER Protein and Germline Transformation Constructs Encoding Wild-Type and Mutant PER Proteins.
(A) Cartoons of the PER structure (amino acids 1–1,224) depicting various functional domains identified by in vitro approaches (upper panel) and of the fragment used for crystallization (amino acids 232–599, lower panel).
(B) 3-D structure of this fragment (adapted from [44]). The approximate positions of the residues Arg345 (red circle), Trp482 (green circle), and Met560 (blue circle) are indicated.
(C) Magnification of the αF:PAS-A interaction surface, where in addition to the mutated residues, the site mutated in perL is indicated.
(D) DNA-constructs encoding wild-type or mutants PER proteins fused to HA or c-MYC tags at their C terminus. Several transgenic lines for each of these constructs where generated (see Materials and Methods). The respective changes are indicated in the same color as in (A). Abbreviations: NLS, nuclear localization domain; PAS, Period-Arnt-Sinlgeminded protein-protein interaction domain; CLD, cytoplasmic localization domain; DBT, Doubletime interaction domain.