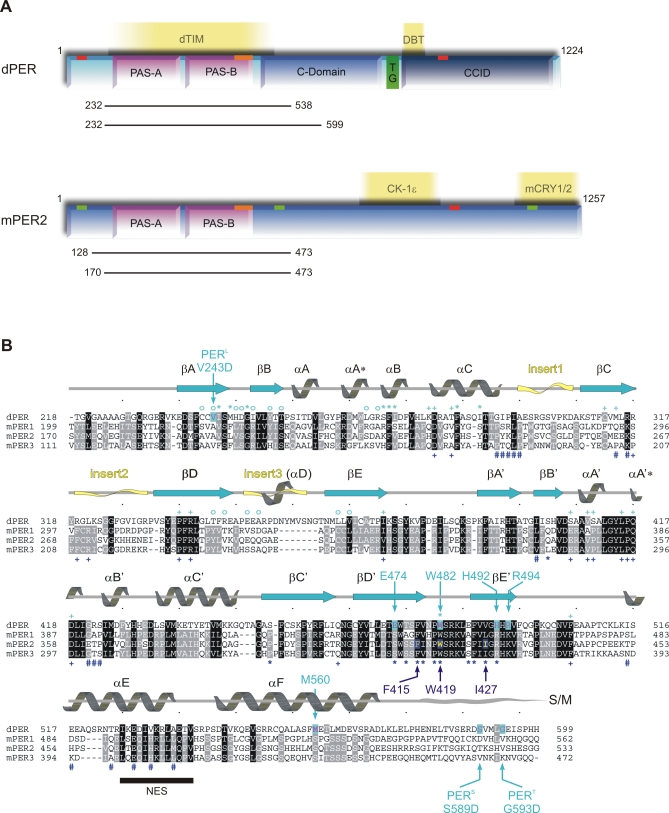
Figure 1. Domain Architecture and Sequence Alignment of dPER and Mouse PERIOD Proteins.
(A) Domain architecture of dPER and mouse PERIOD2 (mPER2). The two PAS domains (PAS-A and PAS-B); the cytoplasmic localization domain (CLD, orange bar); nuclear localization signals (NLS, red bars); NES (green bars); the conserved C-domain; the threonine-glycine (TG) repeat region; and the dCLK:CYC inhibition domain (CCID) of dPER and/or mPER2 are shown schematically. CKIɛ, mCRY1/2, dTIM, and DBT are depicted at their binding sites. The dPER and mPER2 fragments used for crystallization and biochemical studies are represented as black bars.
(B) Sequence alignment of the PAS-A and PAS-B domains as well as helices αE and αF of dPER (Swissprot accession number P07663) with mPER 1, 2, and 3 (Swissprot accession number [1] O35973, [2] O54943, and [3] O70361). Secondary structure elements were assigned from the dPER[232–599] structure ([31], PAS-A, βA → βE; PAS-B, βA′→ βE′). Partially disordered insert regions are depicted as yellow waves. The sequence alignment was generated in ClustalW [78]. dPER residues Trp482, Met560, Glu474, His492, Arg494 mutated herein as well as the sites of the per L long period mutation V243D and the per S/T short period mutations S589D and G593D are highlighted by cyan arrows, mutated mPER2 residues Trp419, Ile427, and Phe415 by dark blue arrows. dPER residues in the Trp482 binding pocket of PAS-A are marked with cyan stars (*), dPER residues interacting with the αF helix with cyan open circles (°). Residues in the dimer interfaces of mPER2 are marked with dark blue stars (*, PAS-B β-sheet interface) or hashes (#, PAS-A dimer interactions). Residues involved in intramolecular PAS domain interactions are marked with cyan (dPER) and dark blue (mPER2) plus signs (+). S/M, short-mutable region.