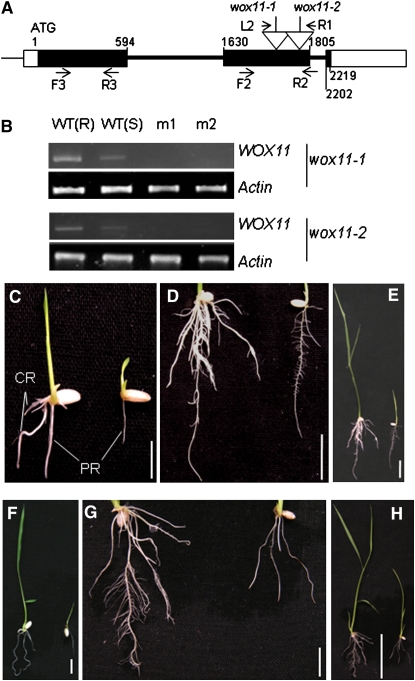
Figure 2.
Characterization of Two T-DNA Insertion Lines of WOX11.
(A) Schematic representation of the WOX11 locus. Relative nucleotide positions of the coding region are indicated (with the initiation ATG codon assessed as 1). The T-DNA insertion positions in the second exon are indicated by open arrows. The positions of the primer L2 and R1 (corresponding to the T-DNA left and right borders, respectively) and the forward (F2) and reverse (R2) WOX11 primers are indicated by arrows. Positions of primers (F3 and R3) used to detect WOX11 expression are indicated. Filled boxes, exons; open boxes, untranslated exons; thick lines, introns.
(B) Detection of WOX11 transcripts in the root [WT(R)] or seedlings [WT(S)] of the wild type or seedlings of wox11-1 and wox11-2. Two different samples (m1 and m2) of each mutant line were used for the RT-PCR analysis. Actin transcripts were amplified as controls.
(C) to (E) Phenotypes of wox11-1 (on the right of each panel) compared with the wild type (left) at 7 d (C) and 14 d ([D] and [E]) after germination.
(F) to (H) Phenotype of wox11-2 (on the right of each panel) compared with the wild type (left) at 7 d (F) and 14 d ([G] and [H]) after germination.
PR, primary root; CR, crown roots. Bars = 1 cm in (C), (D), (F), and (G), 5 cm in (E), and 2 cm in (H).
[See online article for color version of this figure.]