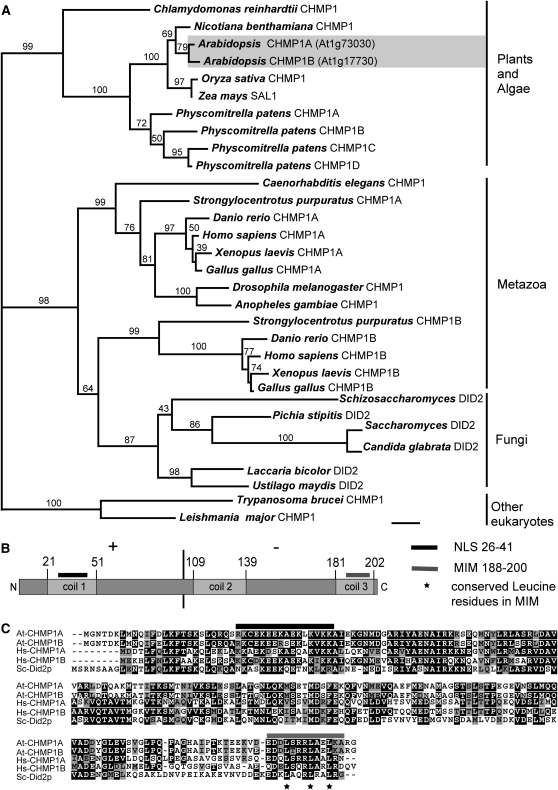
Figure 1.
Characterization of Arabidopsis CHMP1 Protein Structure and Phylogeny of CHMP1 Proteins in Eukaryotes.
(A) Phylogenetic analysis of CHMP1-related proteins from plants, animals, fungi, and other organisms using RAxML. The bootstrap values are shown above each branch. The accession numbers and gene identifiers for the sequences used in this analysis are provided in Methods. Scale indicates 0.1 amino acid substitutions per site.
(B) Schematic representation of Arabidopsis CHMP1A and B proteins. CHMP1A and B differ in 10 amino acid residues from each other. The asymmetric amino acid charge distribution of CHMP1 proteins is indicated by + and − for predominantly basic and acidic amino acid residues, respectively. NLS, nuclear localization signal. Coiled-coil domains were identified with the algorithm from Lupas et al. (1991).
(C) Amino acid alignment of Arabidopsis CHMP1A and B, human CHMP1A and B, and yeast Did2p. Black indicates identical residues, and gray represents similar residues. Asterisks indicate conserved leucine residues in the MIM domain.