Table 2. Crystallographic data.
| Dataset | Native | Seleno-Methionine Derivative |
| Space group/cell constants (a and c, in Å) | P42212/93.74; 113.73 | P42212/93.64; 113.77 |
| Number of measurements/unique reflections | 613,987/47,568 | 320,816/29,379 |
| Resolution range (Å) (outermost shell) | 48.6–1.80 (1.90–1.80) | 48.6–2.11 (2.22–2.11) |
| Completeness (%) | 100.0 (99.9) | 98.4 (89.2) |
| Rr.i.m. ( = Rmeas)/Rp.i.m a , b , c | 0.101(0.696)/0.028(0.190) | 0.081(0.224)/0.027(0.089) |
| Average intensity over st. dev. (<[<I>/σ(<I>)]>) | 23.0 (4.2) | 23.9 (7.3) |
| B-factor (Wilson) (Å2)/average multiplicity | 17.6/12.9 (13.1) | 21.0/10.9 (5.9) |
| Heavy-atom sites used for phasing/fom d | 9/0.68, 0.84 | |
| Resolution range used for refinement (Å) | 48.6–1.80 | |
| Number of reflections used (test set) | 46,847 (720) | |
| Crystallographic Rfactor (free Rfactor)e | 0.160 (0.187) | |
| No. of protein atomsf/solvent molecules/ligands/ions | 2,870/432/2 heme (with Fe3+); 6 glycerols/5 SO4 2− | |
| Rmsd from target values | ||
| bonds (Å)/angles (°) | 0.012/1.31 | |
| bonded B-factors (main chain/side chain) (Å2) | 0.70/2.19 | |
| Average B-factors for protein atoms (Å2) | 14.1 | |
| Main-chain conformational angle analysis for residues in favored regions/outliers/all residues | 349/0/360 |
Friedel mates were treated as independent reflections in the derivative dataset.
Values in parentheses refer to the outermost resolution shell.
 and
and 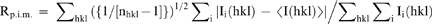 .
.
Mean figure or merit computed for data to 1.8 Å before and after density modification with program DM within CCP4.
Crystallographic Rfactor = Σhkl ||Fobs| − k |Fcalc||/Σhkl |Fobs|; free Rfactor, same for a test set of reflections not used during refinement.
Including atoms in alternate conformation.
