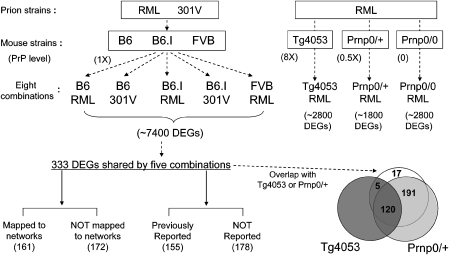
Figure 1.
Strategies for identification of 333 core differentially expressed genes (DEGs) and their functional analysis in mouse prion diseases. Two prion strains (RML and 301V) were used for inoculating mice from six different genetic backgrounds (B6, B6.I, FVB, Tg4053, 0/+, and 0/0) to generate eight prion–mouse combinations. From the list of 7400 DEGs identified from at least one of the five combinations with normal levels of prion protein (1X), 333 DEGs shared by all five were selected through novel statistical methods to represent perturbed networks essential to prion pathophysiology. Venn diagram shows the overlap of the 333 DEGs with DEGs from Tg4053-RML (mice expressing eight times of normal prion protein levels) and from 0/+-RML (mice expressing one-half of normal prion protein levels). Among 333 DEGs, 161 genes were mapped to networks through protein–protein interaction network or metabolic pathways. Also, by comparison of 333 DEGs with previous prion microarray studies, we identified 178 DEGs that have not been reported in connection with prion disease.