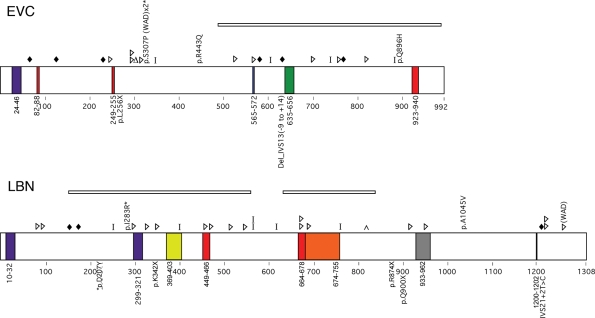
Figure 1.
Summary of expected EVC and LBN protein changes based on previously reported mutation data (top of protein) or those identified in this study (bottom of protein) for patients with EvC syndrome. Colored boxes indicate putative domains. Purple, transmembrane; Red, nuclear localization signal; Green, leucine zipper; Blue, ATP/GTP binding site; Yellow, Flagellar hook associated protein; Orange, Helix-loop-helix; Gray, IQ motif; Black, RGD Cell attachment sequence. Shapes indicate predicted protein changes. Diamonds, splicing error; Triangles, insertion or deletion resulting in a frameshift; Delta sign, deletion of a single amino acid; Bar, truncation mutation; Caret, duplication. Deletions are indicated by rectangles over the deleted area. Asterisks denote missense mutations that occur in an amino acid conserved in mouse and chicken. WAD, Weyers Acrodental Dysostosis.