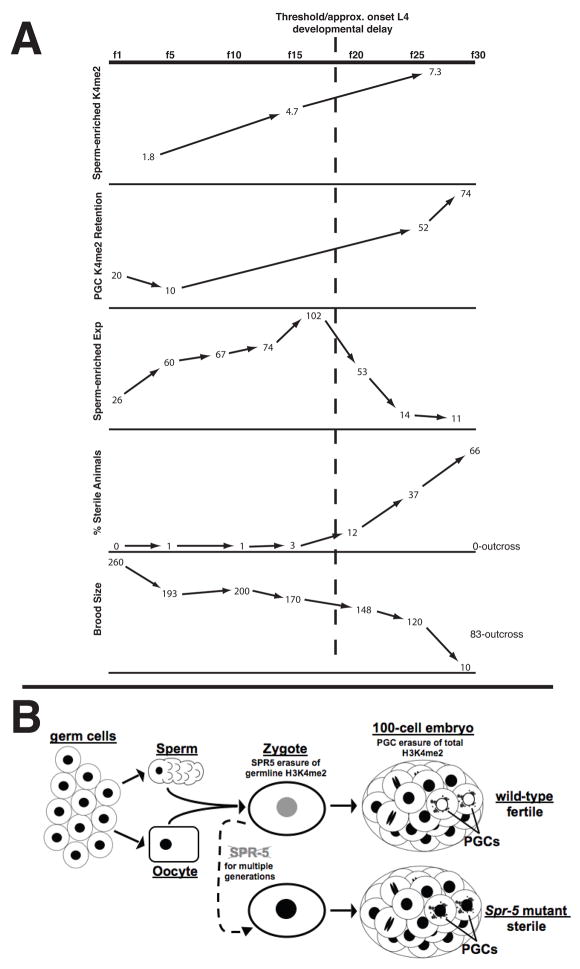
Figure 7.
The summary of data (A) and model (B). A, Graphs comparing the percentage of H3K4me2 precipitated by ChIP in spermatogenesis-expressed genes(Figure 6D), the percentage of H3K4me2 retention in the PGCs (Figure 4), the relative expression of spermatogenesis-enriched genes (Figure 6C), the percentage of sterile animals (Figure 2B) and the brood size (Figure 2A) over generations in spr-5 mutants. The trans-generational increase in H3K4me2 in spermatogenesis-enriched genes due to the failure to demethylate results in increased H3K4me2 retention in the PGCs and the continuous misexpression of spermatogenesis-enriched genes. This leads to an increasing percentage of sterile animals and decreasing brood size, which can be reset by outcrossing the mutation (Figure 2). B, Germ cells, sperm and oocytes have high levels (black nuclei) of H3K4me2 throughout the genome. SPR-5 erases the epigenetic memory of germline transcription by demethylating H3K4me2 at germline-expressed loci in the gametes or early embryo (grey nucleus). A second mechanism removes H3K4me2 throughout the genome in the PGCs (white nuclei). In spr-5 mutants, the failure to reset germline H3K4me2 over multiple generations results in the inappropriate retention of H3K4me2 in the PGCs (black nuclei) and sterility.