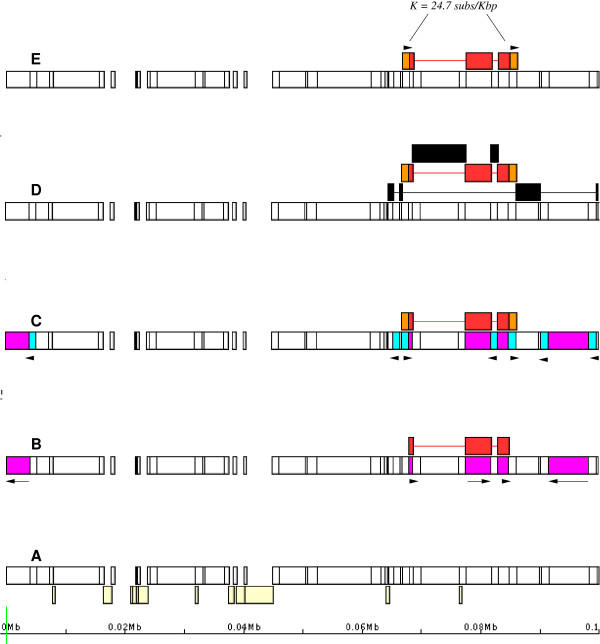
Figure 1.
Re-annotation algorithm. A. Graphic representation of the input REPEATMASKER annotation of the first 100 Kb of genomic sequence [GENBANK:AF123535.1] around the adh gene of the maize cultivar LH_82 (this refers to the same maize sequence that was manually annotated in [45] and used to validate REANNOTATE's predictions in Results, Table 1, and Figure 3). B. Boxes highlighted in magenta on the bottom tier represent hits to the reference element PREM2_ZM_I (the internal region of an LTR-retrotransposon in REPBASE), of which the three innermost hits, shown again in red on the top tier united by a horizontal line, were defragmented by REANNOTATE into a repetitive element model. The black arrows show the orientation of the hits on the chromosome, and the three hits shown in red are colinear with the reference PREM2_ZM_I sequence. C. Boxes highlighted in blue on the bottom tier represent hits to the reference LTR sequence PREM2_ZM_LTR (in REPBASE). Above, two (single-hit) LTR models (shown in orange) flank an IR model (in red): these three models have been assembled into a higher-order model of an element of the PREM2_ZM family. D. The chromosomal span of the defragmented PREM2_ZM element (red and orange) is within the span of another element (bottom model in black); the PREM2_ZM element is inferred to have inserted into the element shown below it. Two other elements (black boxes on top tier) are inferred to have inserted into the PREM2_ZM element. E. Pairs of intra-element LTR sequences are output, aligned with CLUSTALW, and the number of point substitutions between them estimated.