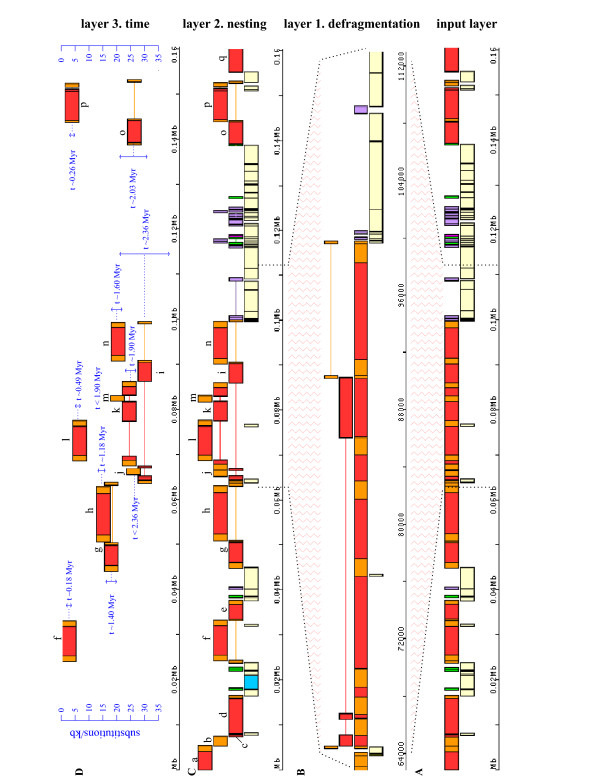
Figure 3.
Layers of re-annotation: maize adh1 locus. A. Representation of the input REPEATMASKER annotation of 160 Kb of sequence around the adh1 locus of maize cultivar LH82. In the bottom tier pale yellow boxes correspond to un-masked sequence (no similarity to known repeats), dark vertical lines indicate low-complexity repeats. Boxes in top tier represent similarity hits to dispersed repeats, separated by vertical lines. Red boxes represent hits to the IR of LTR-elements, orange boxes LTRs, lilac boxes non-LTR retrotransposons, dark green boxes DNA transposons, and the pink box an unknown type of repeat. B. Detail of re-annotation layer 1: Defragmentation. The two bottom tiers represent a portion of the REPEATMASKER annotation shown in A, whilst boxes above indicate 3 IR hits (third tier from bottom) and 3 LTR hits (top tier) defragmented into a single model, and therefore inferred to share an insertion event. This element is labeled 'i' in C and in Table 1, and corresponds to element Victim in 45. (Three hits modeled as part of the same IR are linked by a red line, two hits modeled as part of the second LTR linked by an orange line). C. Re-annotation layer 2: Nesting Structure. Overlapping element models are shown in their order of insertion as resolved by REANNOTATE. (Figure created by rendering in APOLLO GFF annotation automatically generated by REANNOTATE). Letters label LTR-elements in Table 1 that were annotated in 45. D. Re-annotation layer 3: Time. For 'complete' LTR-elements the number of substitutions (vertical scale) between intra-element LTRs and the time since insertion have been automatically estimated. Upper bounds on the ages of two solo LTRs (labeled 'j' and 'm') could be placed as the elements are inserted into complete LTR-elements. Double-headed arrows span two standard deviations around estimates of K. This figure may be compared to Figures 1 and 2 in 45.