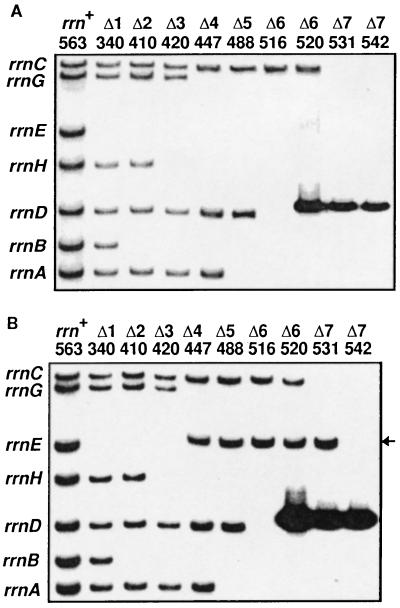
Figure 3.
Autoradiograms of Southern blot hybridization showing the inactivation of chromosomal rRNA operons. DNA was analyzed as described in ref. 26. The number of operons inactivated and strain numbers are shown on the top. The rRNA operon carried by each fragment is shown on the left. (A) Inactivation of the 16S rRNA genes. The SalI–SmaI fragment (probe I, Fig. 1A) of the 16S rRNA gene was labeled with 32P and used for hybridization. TA520, 531, and 542 contain an rRNA plasmid, pHK-rrnC+ and thus give an additional band (8.4 kb) carrying the plasmid-borne rrnC operon (see Fig. 1C) just above the rrnD band (8.1 kb). (B) Inactivation of the 23S rRNA genes. Probe I was removed from the membrane shown in A, and the cellular DNA was rehybridized with 32P-labeled probe II carrying the DNA sequence between the HpaI and the SalI sites in the 23S rRNA gene (Fig. 1A). TA520, 531, and 542 again gave a plasmid-derived band. The ΔrrnG∷lacZ+ construct contains the HpaI–SalI region of the gene (Fig. 1A). Therefore, in TA447, 488, 516, 520, and 531 in which the rrnG operon was inactivated with this construct, the rrnG+-containing band (15.5 kb) disappeared and a new band with the expected size (11.6 kb, indicated by an arrow) appeared just above the rrnE band (11.2 kb). This band disappeared in TA542 in which ΔrrnG∷lacZ+ was replaced with ΔrrnG∷cat+.